Authors
Affan Shoukat1, PhD, Pratha Sah1, PhD, Abhishek Pandey1,PhD, Chad R Wells1, PhD, Yaning Wang2, PhD, Zheng Wang3, PhD, Burton H. Singer4, PhD, Alison P. Galvani1, PhD, Seyed M. Moghadas5, PhD
1 Center for Infectious Disease Modeling and Analysis (CIDMA), Yale School of Public Health, New Haven 06510 USA
2 State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China 100101
3 Department of Biostatistics, Yale School of Public Health, New Haven 06510 USA
4 Emerging Pathogens Institute, University of Florida, Gainesville, FL 32610, USA
5 Agent-Based Modelling Laboratory, York University, Toronto, Ontario, M3J 1P3 Canada
Abstract
In December 2019, a novel coronavirus (SARS-CoV-2) was identified from a cluster of patients with severe pneumonia-like symptoms in Wuhan, China. The rapid spread of disease (COVID-19) in China and importation of cases to other countries has led to a global public health emergency, with concerns over its pandemic potential. Understanding the epidemiological and transmission characteristics of COVID-19 is essential to prevent and mitigate future outbreaks.
We quantified the transmissibility (i.e., the basic reproduction number, R0) of the virus from the initial cluster of COVID-19 patients. We used symptom onset dates of 41 cases from December 1, 2019 to January 1, 2020 and estimated mean R0 to be 4.1 with a quantile range of 1.7 – 7.6 on December 15. Our larger R0 estimate compared to previous studies indicates the potential for substantial human-to-human transmission in the initial stages of the Wuhan outbreak, highlighting the importance of rapid identification of symptomatic patients, self-isolation, and quarantine for probable cases in the absence of vaccination.
Keywords
COVID-19, transmissibility, reproduction number
REPORT
Since being identified as a novel coronavirus (SARS-CoV-2) outbreak in Wuhan city, over 74,000 cases and 2,100 deaths in China have been reported as of February 20, 2020, of which more than 60,000 cases are from Hubei Province, the source of infection (1). Additional cases have been confirmed in several other countries, including the United States, Canada, France, Australia and Japan. The case fatality rate is estimated to be about 2.2% (2), orders of magnitude higher than a typical coronavirus and 20 times higher than a typical influenza A virus (3), the latter of which usually leads to over 300,000 annual deaths around the world (4). The combination of rapid spread and importation of cases to other countries poses a significant global health threat. Without a vaccine or antiviral treatment, control of the virus has relied on screening and travel restrictions. On January 23rd, a lockdown was enacted that banned any travel out of the urban epicentre, and public gatherings for Lunar New Year in Wuhan were cancelled
(5).
At the early phase of an outbreak, it is essential to quantify the transmission characteristics of the disease to determine its potential impact as a public health emergency of international concern (6). As countries around the world strive to prepare for potential outbreaks of COVID-19, this quantification can help decision-makers to identify the type and intensity of control measures required to mitigate infection spread. The basic reproduction number (R0), defined as the average number of secondary cases generated by a single infection in a susceptible population, is a fundamental epidemiological parameter that quantifies the transmissibility of a disease and governs the trajectory of its outbreak.
We sought to estimate R0 based on the earliest dates of clinical identification of COVID-19 patients in the epicentre. We fitted a generalized Richards model (9) to the symptom onset dates of 41 initial cases in Wuhan from December 1, 2019 to January 1, 2020 (10) using a least-squares algorithm. The parameters of fitting were then used to generate incidence curves from negative-binomial distributions, and to forecast the growth of the outbreak. We generated 500 epidemic curves using Monte Carlo iterations, while accounting for observational error with overdispersion for incidence on each calendar day. For calculating R0, the serial interval (defined as the duration between the onset of symptoms in a source case to a secondary case) was sampled from a gamma distribution with mean of 7.5 days and standard deviation of 3.5 days for the COVID-19 (7). Using R0 estimates, we next quantified the reduction in daily contacts of symptomatic patients needed to bring R0 below 1 and avert large outbreaks. We used negative binomial distributions to sample the number of daily contacts for each symptomatic patient (according to an age-specific matrix of contacts for urban settings (11)), over the symptomatic period without any other control measures. The analysis and computational codes are available online at http://github.com/affans/2019-ncov.
We estimated R0 for the initial cluster of COVID-19 patients to have a mean of 4.1 with a quantile range of 1.7 – 7.6 on December 15 (Figure 1A). This corresponds to an infection probability of 0.043 (quantile range 0.018 – 0.08) per contact. A number of studies have to-date estimated R0 using the dates for reported confirmed cases since January 11, 2020, with overlapping ranges and mean values between 2 and 3 (7,8). Our larger estimate of R0 (using symptom onset dates) compared to previous studies indicates a potential for substantial human-to-human transmission in the initial stages of the Wuhan outbreak, and suggests that a higher intensity of interventions may be needed at the outset of disease to prevent its exponential dissemination, especially in urban and densely populated settings. For mean values of R0 in the range 2 – 4.5 from previous studies and ours, we found that the daily number of contacts for symptomatic patients would need to be reduced by 50% – 78% to achieve the threshold of R0 = 1, below which the outbreak is expected to decline and diminish (Figure 1B). Overall, these results indicate that control measures would need to interrupt a substantial level of transmission in order to prevent future, or curb ongoing outbreaks, highlighting the importance of rapid identification of symptomatic patients, self-isolation, and quarantine for probable cases in the absence of vaccination.
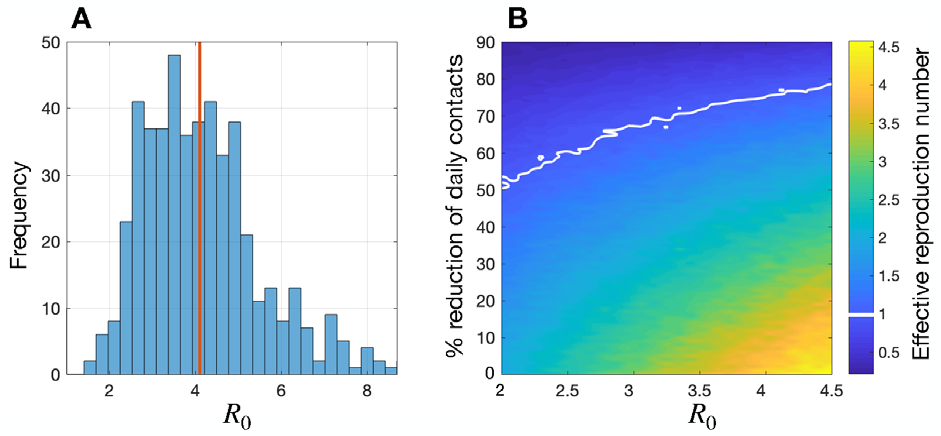
(B) Colorbar represents reduced R0 (i.e., the effective reproduction number) attributed to
the percentage of daily contacts avoided during symptomatic disease for different R0 values.
References
- National Health Commission of the People’s Republic of China. Website [Internet]. [cited 2020 Jan28]. Available from: http://www.nhc.gov.cn/xcs/xxgzbd/gzbd_index.shtml
- Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet [Internet]. 2020 Jan 24; Available from: http://www.sciencedirect.com/science/article/pii/S0140673620301859
- Wong JY, Kelly H, Ip DKM, Wu JT, Leung GM, Cowling BJ. Case fatality risk of influenza A (H1N1pdm09): A systematic review. Epidemiology. 2013 Nov;24(6):830–41.
- WHO | Burden of disease. 2019 Nov 22 [cited 2020 Jan 30]; Available from: https://www.who.int/influenza/surveillance_monitoring/bod/en/
- China halts flights and trains out of Wuhan as WHO extends talks [Internet]. CNA. [cited 2020 Jan 28]. Available from: https://www.channelnewsasia.com/news/asia/wuhan-virus-quarantine-city-flightstrains- china-12306684
- Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV) [Internet]. [cited 2020 Feb 13]. Available from: https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-theinternational- health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novelcoronavirus-( 2019-ncov)
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med [Internet]. 2020 Jan 29; Available from: http://dx.doi.org/10.1056/NEJMoa2001316
- Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet [Internet]. 2020 Jan; Available from: https://linkinghub.elsevier.com/retrieve/pii/S0140673620302609
- Mostaço-Guidolin LC, Greer A, Sander B, Wu J, Moghadas SM. Variability in transmissibility of the 2009 H1N1 pandemic in Canadian communities. BMC Res Notes. 2011 Dec 13;4(1):1–9.
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb;395(10223):497–506.
- Mossong J, Hens N, Jit M, Beutels P, Auranen K, Mikolajczyk R, et al. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 2008 Mar 25;5(3):e74.
Production of this document has been made possible through a financial contribution of the Public Health Agency of Canada to the National Collaborating Centre for Infectious Diseases. The views expressed herein do not necessarily represent the views of the Agency or NCCID.
For more on mathematical modelling and public health:
- Imperial College London: MRC Centre for Global Infectious Disease Analysis – News Items -COVID-19
- Online discussion group – mod4PH
- Mathematical Modelling in Public Health – Video
