COVID-19 Real-Time RT-PCR diagnostic tests can provide information on whether or not a patient has been infected with SARS-CoV-2 by detecting and measuring the virus’ genetic material.
This document provides a brief description of the Real Time Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) test. It provides basic information on how this molecular biology test is used in laboratories to detect genetic material of a pathogen such as SARS-CO-2 virus, the cause of COVID-19 disease. It also provides the basics on interpreting test results that can be helpful in understanding the state of the disease and/or its progression and the likelihood of transmissibility. It can be of value to public health practitioners or anyone in healthcare community involved in the COVID-19 response.
What is a genetic material and how is it used to test for infectious diseases?
Genetic material is the instruction manual within a cell or virus that provides the directions on how to behave, survive and more. There are two types of genetic materials: Deoxyribonucleic acid (DNA) and Ribonucleic acid (RNA). The major difference between the two types of genetic materials is that DNA is a double-stranded structure whereas RNA is single-stranded. In terms of diagnostics, DNA is more stable for testing infectious disease than RNA because of its structural and intrinsic properties. It is also important to note that SARS-CoV-2 contains only RNA.
A common feature among all viruses is that they depend on host proteins and reproductive machinery for survival. Consequently, viruses like SARS-COV-2 are required to invade healthy cells in order to survive and multiply. Similar to other viruses, when SARS-COV-2 infiltrates a cell, it releases its RNA and exploits the cell’s machinery for viral replication. Furthermore, as long as the virus’ genetic material is present inside the cell, we can use a laboratory technique called Real-Time RT-PCR to determine whether a patient has been/is infected with SARS-CoV-2.
What is Real-Time RT-PCR?
Real-Time RT-PCR (Reverse Transcription Polymerase Chain Reaction) is a sensitive and fast test used for detecting the presence of specific genetic materials within a sample. This genetic material can be specific to humans, bacteria, and viruses like SARS-CoV-2.
The foundation of Real-Time RT-PCR derives from Polymerase Chain Reaction (PCR); a laboratory technique developed by Nobel Prize-winner, Kary B. Mullis, in the 1980s, to allow researchers to amplify and detect specific DNA targets (1,2). This technology was later improved to allow “real-time” visualization and quantification of DNA targets as they undergo amplification. To visualize the amplification of DNA, Real-time PCR uses increases in the fluorescence intensity of a fluorogenic probe in proportion to the amount of amplified DNA. By measuring this fluorescence intensity, one can quantify the amount of genetic material inside the sample. A major limitation of PCR is that it detects only DNA templates. Thus, in order to apply Real-Time PCR on RNA samples (i.e. genetic material of SARS-CoV-2), researchers have to use a special enzyme – called Reverse Transcriptase – to convert RNA into DNA templates, also known as complementary DNA (cDNA). Altogether, these features contribute to the versatility and sensitivity of Real-Time RT-PCR as a diagnostic test for infectious diseases.
How does Real-Time RT-PCR Work?
Sample Collection: To start the diagnostic test, a trained healthcare worker will use a swab to collect nasopharyngeal specimens from the patient’s
nasopharynx. The sample is then placed into a sterile tube containing viral transport media to keep the virus viable (3).
Sample Preparation: Once the specimens arrive at the laboratory, researchers will use available commercial purification kits to extract RNA from the sample. Next, the RNA sample is added into one reaction mixture containing all the ingredients required to complete the diagnostic test, also known as “one-step RT-PCR”. The ingredients inside this mixture includes DNA polymerase, reverse transcriptase, DNA building blocks, and specific fluorophore probes and primers that recognize SARS-CoV-2.
Reverse Transcription: As mentioned earlier, PCR only works on DNA templates. Thus, the role of reverse transcriptase inside the reaction mixture is to convert all the RNA present within a given sample into cDNA. This includes human RNA, bacterial RNA, even other coronavirus RNA and if present, SARS-CoV-2’s RNA.
Step 1 – Denaturation/Separation: To begin, it is important to remember that DNA is a double-stranded structure. Thus, the next step is to unwind the DNA molecule into separate DNA stands. This is accomplished by heating the DNA to high temperatures (> 90°C) for amount 10 min.
Step 2 – Primer Annealing: Next, is the addition of short fragments of DNA, called primers. Primers are designed with high specificity and will attach to specific targets within cDNA of the SARS-CoV-2 RNA virus. The specific lower temperature is needed for primer annealing too. In general, there 7 common gene targets used for testing COVID-19; each gene target is essential to the virus’ replication or structure(4). Those essential gene targets include RNA-dependent polymerase (RdRP), ORF1ab (SARS-CoV-2’s conserved open reading frame), S gene (spike protein), N gene (nucleocapsid protein), E gene (envelope; virus’ outer shell).
Step 3 – Primer Extension/Elongation: Since DNA is a double stranded structure, there are two primers in this reaction mix, each one is designed to target one of the two DNA strands. Once the primers attach to their target DNAs, they will direct the DNA polyermase on where to begin and finish amplification on the DNA segment. This step results in an identical DNA copy of the target DNA.
And Repeat: Real-Time PCR will repeat the cycle multiple times (usually for 40 cycles). Every time RT-PCR completes a cycle, it will double the target DNA. Additionally, there are also fluorescent probes that bind specifically to the DNA targets, downstream of each primer. Every time DNA polyermase amplifies the DNA target, it will activate the probe to release a fluorescence signal. Thus, as the amount of target DNA increases, the fluorescence intensity will also increase.
What is the readout of Real-time RT-PCR?
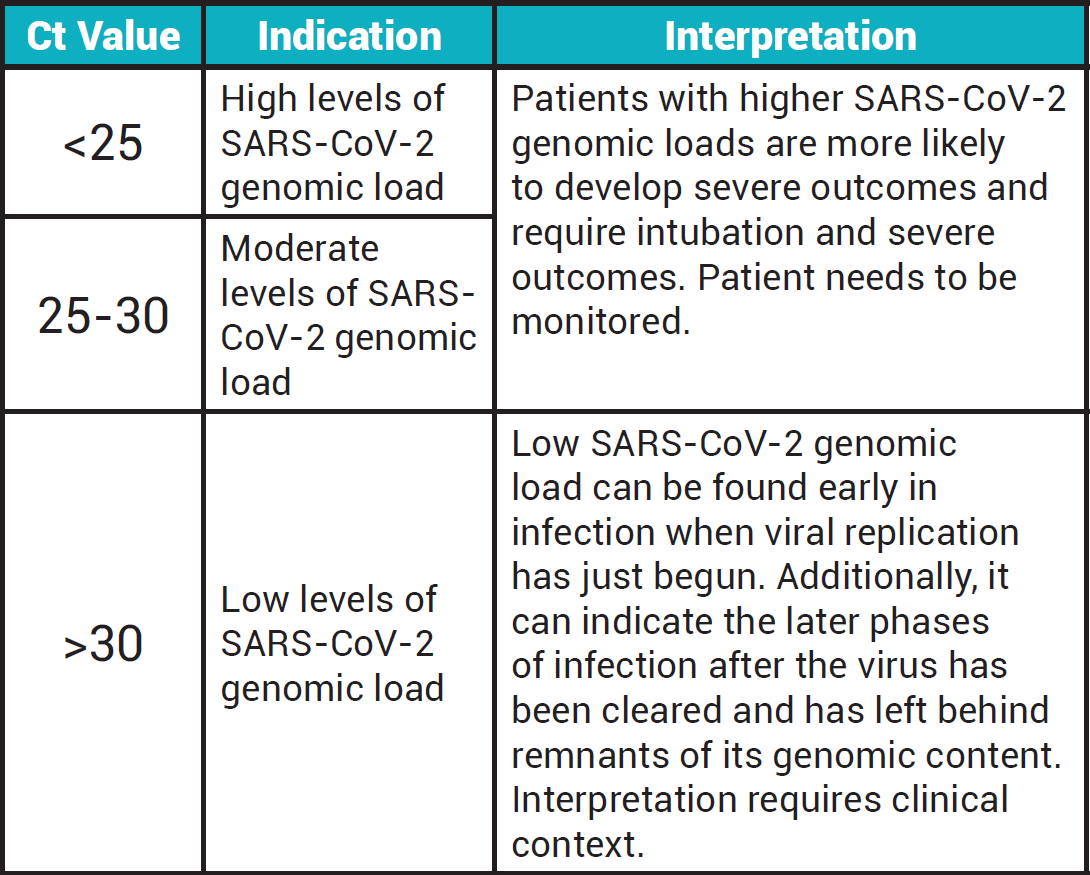
The emitted fluorescence is then captured as a signal to generate a ‘cycle threshold’ (Ct) value. The Ct values refers to the amount of cycles required for the fluorescent signal to exceed background levels. Generally, the more target DNA that is in the sample, the faster its amplification will be and thus, the fewer cycles required before the fluorescence signal crosses the background threshold (lower Ct value). Conversely, if there are low amounts of target DNA, it will require more cycles before the fluorescence can cross the background threshold (higher Ct value).
Why are Ct values important?
Ct values are useful because they provide information about the patient’s pathogen genetic material load (SARS-CoV-2). A low Ct value indicates high viral genomic load, whereas a high Ct value indicates low viral genomic load. Health professionals can use Ct values in conjunction with clinical symptoms and history to gauge a patient’s stage of disease. Furthermore, serial Ct values generated from repeated testing can also help clinicians monitor the disease progression and predict stages of recovery and infection resolution. Contact tracers also utilizes Ct values to prioritize their attention to patients with the highest viral genomic load, which indicates a high risk for transmissibility.