Publisher’s Note
On behalf of the National Collaborating Centre for Infectious Diseases, I am pleased to introduce the Surveillance of Antimicrobial Resistance and Antimicrobial Utilization in Canada report. We commend its authors and their multidisciplinary project team for their work in addressing this important subject. We are confident that this report can serve at least three purposes. First, it informs public health decision-makers and other stakeholders of past and recent achievements and the current status of surveillance in Canada. Second, it provides comparative information with respect to surveillance systems in other countries. Third, it identifies strengths and weaknesses of surveillance today as well as opportunities for further improvements. This report and the recommendations of the authors can be found by visiting the AMMI website at www.ammi.ca.
Joel Kettner
Scientific Director
NCCID
Preface
Antimicrobial Resistance in Canada – Paused at a Crossroads
This report, on the current status and future possibilities for antimicrobial resistance and utilization surveillance in Canada, was commissioned by the National Collaborating Centre for Infectious Diseases. Our group, based in the Association of Medical Microbiology Infectious Disease Canada, drew extensively on the expertise of many colleagues involved in animal, human, and public health across the country.
Many countries have completed or are in the midst of similar reviews of surveillance. Why? The threat of antimicrobial resistance has been understood since the dawn of the antibiotic era. Sir Alexander Fleming, accepting the 1945 Nobel prize for the discovery of penicillin, presciently sounded a warning note about resistance in the event of improper penicillin use.1 Over the years, as common bacterial pathogens evolved the genetic arsenals that now allow them to withstand our antibiotics, we kept a step ahead by developing new antimicrobials, and thus maintained the remarkable medical miracle of antibiotic therapy.
However, in recent years, increasing resistance rates and new, quickly spreading and highly resistant microbes, in concert with reduced new antimicrobial development, have rightfully raised a global alarm.2 Within the past year, the U.S. Centers for Disease Control and Prevention, the European Centre for Disease Prevention and Control, the World Health Organization, and the World Economic Forum have all released reports or held meetings addressing the dangers of antimicrobial resistance (AMR).3-6 The latter group, in its 2013 annual report on global risks concluded “arguably the greatest risk … to human health comes in the form of antibiotic-resistant bacteria. We live in a bacterial world where we will never be able to stay ahead of the mutation curve. A test of our resilience is how far behind the curve we allow ourselves to fall.”6
The World Health Organization has made AMR a priority, and has issued centralized direction for member nations. The key messages from an AMR group at the 2013 World Health Assembly concisely stated: “Antibiotic resistance is a rapidly evolving health issue extending far beyond the human health sector. Awareness of the seriousness of the situation and the need for urgent action is required at the highest political level, globally and at country level. A cross-sectoral approach including agriculture, fisheries, development and economics is required for effective action at global and national levels.”
In Canada, establishing effective surveillance of AMR with its cause and counterpart, antimicrobial utilization has been hampered, perhaps by the very nature of our nation: a geographically vast, culturally diverse, and medically advanced nation, with multiple jurisdictions of public health and government. To be able to address this public health problem, however, we must understand its scope and actively monitor its spread. Gaps in our current surveillance are clear. We have some sound elements that address aspects of the needed surveillance, but we lag behind many international counterparts. We need to align and combine our existing provincial and federal surveillance resources—which fortunately are many—for cohesive, integrated, accountable national surveillance that can evolve and expand to meet the AMR challenge. At its core, public health surveillance is information for action. It is the foundation we need to develop our response to this threat to modern medicine, preserve the precious resource that is antibiotic therapy, and protect the health of Canadians now and in the future.
Dr. Lynora Saxinger
Chair, Antimicrobial Stewardship and Resistance Committee
Association of Medical Microbiology and Infectious Disease Canada
Executive Summary
This report summarizes the results of a 2012–2013 project sponsored by the National Collaborating Centre for Infectious Diseases. The task set forth was both to assess the current status of surveillance of antibiotic or antimicrobial use (AMU) and antimicrobial resistance (AMR) in Canada, and to provide recommendations to advance Canadian surveillance.
Both ‘antimicrobial’ and ‘antibiotic’ utilization are terms in common use, but the broader term, antimicrobial, will be preferred in this report. It is implicitly understood throughout this report that the surveillance of antimicrobial resistance as a public health threat must be accompanied by surveillance of its main modifiable driver, antibiotic utilization.
This project had three main components:
1. A systematic literature search (Appendix A) was performed to identify, describe and evaluate Canadian and international AMR and AMU surveillance programs, with analysis of their attributes. A structured evaluation method was applied to exemplar Canadian and international systems, and more a detailed analysis review of two models of surveillance (Denmark’s DANMAP program and the European Centre for Disease Control and Prevention’s EARS-Net and ESAC-Net programs) was performed to identify their relevance, strengths and weaknesses in potential application to a Canadian context.
2. A semi-structured interview protocol (Appendix C) surveyed Canadian experts from key stakeholder groups (including but not limited to public health and infectious diseases physicians, physician and PhD microbiologists, antimicrobial pharmacists, veterinarians, and representatives of the food animal industry) to ensure a full understanding of current functioning Canadian AMR and AMU surveillance in all sectors, to identify perceived strengths, and weaknesses, and to identify perceived needs.
3. A review of previous Canadian antimicrobial resistance and surveillance consensus meetings, reports and recommendations to inform the development of an actionable set of recommendations (Appendix E).
The findings of each component were complementary in evaluating the past, present, and potential future of AMR and AMU surveillance in Canada. The systematic review of 20 databases and grey literature reviewed 8931 studies, selected 129 studies for analysis, and identified 11 Canadian and 58 international surveillance programs and projects, including all programs identified in the expert surveys. Review of the major Canadian programs and detailed comparison with select international programs revealed that in spite of focal high quality surveillance components, Canadian public health AMR and AMU surveillance can be seen as having significant gaps, incomplete integration, with no one defined accountable body responsible for surveillance, and no single national mandate. Specific shortcomings in the scope of current Canadian AMR and AMU surveillance programs include: constricted focus (by pathogen or population), a lack of public-health based data on evolving community and hospital AMR (beyond antibiotic resistant organisms of interest to infection control), and challenging but improving access to antibiotic utilization data to inform development of antimicrobial stewardship in human and agri-food/veterinary populations. The most significant gap in surveillance data that can be immediately addressed is community-based AMR surveillance, as all extant published surveillance data are from non-public-health collations of hospital microbiology laboratory bacterial susceptibility data. In identifying a way forward, we noted that the most highly evaluated international public health surveillance programs, despite different structures, have integrated oversight of national and regional surveillance, across veterinary, food animal and human medicine (that is, with food agency and public health agency collaboration). We conclude that the complex ecology of antimicrobial resistance development requires a cross-sectoral, trans-disciplinary, integrated approach for appropriate surveillance to inform control efforts.
Finally, the review of Canadian reports and conferences (including the relevant reports of the Auditor General of Canada) addressing antimicrobial resistance and surveillance over the last 17 years allowed an assessment of their impact and identified challenges that may interfere with future progress (see Appendix E for summarized recommendations). The reports range from the 1997 Controlling Antimicrobial Resistance: An Integrated Action Plan for Canadians, through to the most recent Canadian Committee on Antimicrobial Resistance (CCAR, now disbanded) reports of 2004 and 2009. Repeated recommendations from an increasing range of sources portray a history of consistently known goals that have not yet been met. A review of the reports of the Auditor General of Canada pertaining to Health Canada’s (and later PHAC’s) surveillance activities suggests that the lack of an effective provincial-territorial-federal agreement structure and surveillance framework is relevant to the lack of progress observed. Increasingly specific calls have been made for national, public health-based coordination of true national AMR and AMU surveillance. We contend that the protection of public health from consequences of antimicrobial resistance in microorganisms is a shared responsibility including federal leadership for a strong national, public health based coordination of surveillance, with engagement and collaboration of provincial and territorial health agencies, professional associations, animal health, and food animal-industry stakeholders. It is our hope that this report appropriately builds on the work of predecessors, integrates the valuable experience of international colleagues, and will help establish the structure, collaborations, and momentum required to appropriately track (and therefore permit us to address) antimicrobial resistance as an evolving public health threat in Canada.
Principal Investigators
Dr. Lynora Saxinger, Dr. Jennifer Grant, Dr. David Patrick
With our colleagues in the Project Steering Committee:
Patrick Boerlin DVM
William Bowie MD FRCPC
Elizabeth Bryce MD FRCPC
Carolee Carson DVM PhD
John Conly MD FRCPC
Patricia Dowling DVM MSc DACVIM DACVCP
Kevin Forward MD FRCPC
Denise Gravel BScN MSc PhD (c) Jim Hutchison BSc. MD FRCPC
Tim Lau BSc PharmD FCSHP
Fawziah Marra PharmD FCSHP
Andrew Morris MD FRCPC SM
Lindsay Nicolle MD FRCPC
Richard Reid-Smith DVM
Craig Stephen DVM PhD
Karl Weiss MD MSc FRCPC FR
List of Acronyms, Abbreviations and Definitions
ADD Defined animal daily doses
AMR Antimicrobial resistance
AMU Antimicrobial use
API Active pharmaceutical ingredient ARO Antibiotic resistant organism(s)
BDs Bed days
CANWARD Canadian Ward Surveillance Study
CBSN Canadian Bacterial Surveillance Network
CCAR Canadian Committee on Antibiotic Resistance
CDAD Clostridium difficile-associated diarrhea
CFIA Canadian Food Inspection Agency
CIPARS Canadian Integrated Program for Antimicrobial Resistance Surveillance
CHEC Canadian Hospital Epidemiology Committee
CLSI Clinical and Laboratory Standards Institute
CNISP Canadian Nosocomial Infection Surveillance Program
CRE Carbapenem-resistant Enterobacteriaceae
DDD Daily defined dose
DID DDD (daily defined dose) per 1000 inhabitants per day
ESBL Extended spectrum beta-lactamases
GRADE Grading of Recommendations Assessment, Development and Evaluation
HAI Hospital-acquired infections
ICU Intensive care units
IMS IMS Health Inc.
MIC Minimum inhibitory concentration
MRSA Methicillin-resistant Staphylococcus aureus
NGO Non-governmental organization
NSERC National Sciences and Engineering Council of Canada
OTC Over the counter
PHAC Public Health Agency of Canada
PID Packages per 1000 inhabitants per day
SIR Susceptible, intermediate susceptible, resistant
VRE Vancomycin-Resistant Enterococci
Acknowledgments
The authors thank Diana Kao, Mimi Doyle-Waters and Deirdre Doyle-Waters for their participation in the research conducted for this project. We gratefully acknowledge the anonymous participants who agreed to complete our survey interviews. We wish to express our gratitude to the members of the project advisory committee for their contributions to the design and interpretation of the research, development of the recommendations and technical review of this report:
Patrick Boerlin, DVM
William Bowie, MD, FRCPC
Elizabeth Bryce, MD, FRCPC
Carolee Carson, DVM PhD
John Conly, MD, FRCPC
Patricia Dowling, DVM, MSc, DACVIM, DACVCP
Kevin Forward, MD, FRCPC
Denise Gravel, BScN, MSc., PhD (c)
Jim Hutchison, BSc., MD, FRCPC
Tim Lau, BSc, PharmD, FCSHP
Fawziah Marra, PharmD, FCSHP
Rachel McKay, MSc
Andrew Morris, MD, FRCPC, SM
Lindsay Nicolle, MD, FRCPC
Richard Reid-Smith, DVM
Craig Stephen, DVM, PhD
Karl Weiss, MD, MSc, FRCPC, FRCP
1. Introduction
• This report aims to explore the current systems of surveillance of antimicrobial resistance and antibiotic utilization in Canada and abroad, and to make recommendations for improving surveillance programs in Canada.
• Increasing occurrence of antimicrobial resistance in bacteria is a global public health risk.
• Changes to systems for the surveillance of antimicrobial resistance and antimicrobial utilization (AMR-AMU) in Canada could be improved based on the review presented here as well as on the recommendations from past work.
1.1. Purpose
In the context of the increasing international and national recognition of the imminent threat to public health presented by antimicrobial overuse and resistance, a project to explore ways to optimize surveillance of antibiotic use and antimicrobial resistance in Canada was commissioned by the National Collaborating Centre for Infectious Diseases. Specifically, the project was designed to meet the following objectives:
• To determine core elements of antibiotic use and AMR surveillance initiatives worldwide through systematic search and review of published and grey literature related to surveillance of antibiotic use and antimicrobial resistance in human and veterinary medicine.
• To summarize current national, provincial, and regional AMR surveillance programs and initiatives as well as current national, provincial, regional, and private antimicrobial use monitoring programs/initiatives in Canada.
• To prepare a comprehensive list of existing regional, provincial and national surveillance programs summarizing details of which data are collected, for how long they have been collected, and the specific definitions used for data collection and reports resulting from these data.
• To identify missing elements in existing surveillance programs in Canada and to describe the barriers to the operation of an ideal program through expert reviews and key informant interviews.
• To provide recommendations for implementing optimal antimicrobial use monitoring and resistance surveillance programs in Canada.
1.2. Background
Bacteria have perfected adaptive strategies for species survival for about three billion years, including the development of antimicrobial compounds and resistance to those compounds as part of inter-microbial battles for dominance. Strains isolated from Siberian permafrost sediments dating back millions of years have demonstrated multidrug resistance to tetracycline, streptomycin and chloramphenicol—all antibiotics that are commonly used in medical therapy today.7 Resistance genes developed to allow bacteria to cope with ever-changing biophysical, chemical and ecological conditions of their habitat and are involved in a variety of processes including detoxification, metabolic function and signal trafficking.8 All forms of precursors to resistance elements including bacteria and genes make up the “resistome,” and we now appreciate that the mechanisms that allow resistance determinants to be transferred to human pathogens after exposure to antibiotics were present long before broad therapeutic use.8,9
However, with their intensive use of antibiotic drugs typically administered to patients since the dawn of the clinical antibiotic era has led to selection of some pathogens that are unaffected by them. Well-known pathogens that were previously easily treatable are demonstrating concerning trends of developing antimicrobial resistance, in some cases leading to re-emergence. For example, Canadian surveillance of susceptibilities to Neisseria gonorrhoeae has been systematically collected as part of Public Health Sexually Transmitted Infection surveillance. Data from 2000–2009 have demonstrated that there has been a significant increase in antimicrobial resistance among N. gonorrhoeae isolates to a point where therapeutic use of quinolones is no longer an effective treatment option,10 prompting a change in the national treatment guidelines and demonstrating the importance of resistance surveillance.
As the effectiveness of antibiotics against certain pathogens declines, the morbidity and mortality in human patients as a result of these infections increases. Moreover, the health care costs associated with antibiotic resistant infections also increases.11,12
Among contemporary global issues, the public health risk associated with the development of antimicrobial resistance in clinically important bacteria species has carved a distinct share of concern in the minds of health professionals, policy-makers, and citizens at large. In 2013, the Chief Medical Officer for England, Dame Sally Davies, stated: “Antimicrobial resistance is a ticking time-bomb not only for the U.K. but also for the world. We need to work with everyone to ensure the apocalyptic scenario of widespread antimicrobial resistance does not become a reality. This threat is arguably as important as climate change.” The European Commission presented its first action plan to tackle AMR on November 17, 2011, noting that some 25,000 patients die each year in the European Union from infectious caused by drug-resistant bacteria. A 2013 AMR risk stratification report published by the United States Centers for Disease Control and Prevention (CDC) estimated that at least 23,000 Americans die from antibiotic resistant infections. Although estimates are highly variable, $20 billion in excess direct healthcare costs are linked to antibiotic-resistant illness.3 There is widespread agreement among the scientific, medical and public health community that the time to address this health risk is now.
Antibiotic use is not restricted to human populations. In animals, antimicrobials are used for therapeutic treatment (treatment of animals with a bacterial infection), prophylaxis (treatment of animals to prevent the development of a bacterial infection), metaphylaxis (use of antibiotics to decrease the chances of illness) and antimicrobial growth promotion (use of antibiotics to increase the tonnage of meat produced, unrelated to illness).13 The precise amount of antimicrobial agents used in animal populations is unknown, with some estimates suggesting that agricultural use alone comprises about 80% of overall use by weight of drug.
Veterinary use of antimicrobial agents that are used in human medicine or have a human analogue increases the likelihood that human bacterial pathogens with food animal reservoirs will develop resistance or cross-resistance to antibiotics that are approved for use in human medicine.14 Zoonotic transfer of antimicrobial-resistant pathogens from animals to humans via meat products is well established.15 Although some antimicrobial agents used in animals belong to classes that do not have counterparts in human medicine, commonly used antibiotics, including the tetracyclines, penicillins, macrolides and sulphonamides, are frequently used in both human and veterinary medicine.13 In addition to the impact on human health, antimicrobial resistance in animal pathogens has a deleterious effect on animal health, including substantial economic impact on the food-production sector.
The newest global AMR challenge, carbapenemase-producing enterobacteriaceae, provides a wake-up call to the potential of AMR organisms to spread globally in a short period of time and seriously challenge the capabilities of modern medical therapy. These bacteria, which are resistant to most, and sometimes all, available antibiotics, also illustrate the pervasive nature of AMR organisms in the environment, and the need to monitor resistance outside human health settings as well as within. Carbapenemasecarrying bacteria have been found in non-human sources (although it is unclear if the resistant bacteria were derived from human sources), and can extensively contaminate the environment—especially in settings with poor sanitation. One study revealed carbapenemase-carrying bacteria in multiple water samples from the streets of New Delhi.16 As carbapenems are our last good defence against many resistant Gram-negative bacteria, the need to limit the rise and spread of these bacteria is urgent. Active surveillance in health care, as well as in the food chain and other non-human sources has been emphasized. Until the complex ecology of resistance in human and animal health, food production, and the environment is better understood, ongoing monitoring in all of these spheres remains crucial to allow development of control efforts.
Acknowledging the inextricable interconnection between human health, animal health and overall ecosystem health, the One Health Initiative is a worldwide strategy intended to build a stronger bond between human medicine and veterinary medicine.17 Recent reports from the One Health Initiative highlight the importance of adopting this approach to addressing the public health risk linked to antimicrobial resistance in microorganisms stating that, “there is a significant need to coordinate surveillance efforts at a global scale”.18 Increasingly, the One Health approach to formulating policies and actions is being promoted to facilitate early detection of new diseases that emerge from animal and insect reservoirs, and also to offer potential means for improving food safety and preventing the emergence of antimicrobial resistance in humans and animals.
The World Health Organization defines public health surveillance as the continuous, systematic collection, analysis and interpretation of health-related data needed for the planning, implementation, and evaluation of public health practice. Such surveillance can:
• serve as an early warning system for impending public health emergencies;
• document the impact of an intervention, or track progress towards specified goals;
• monitor and clarify the epidemiology of health problems, to allow priorities to be set and to inform public health policy and strategies.19
The World Health Organization outlines five key advantages for establishing a national or regional surveillance network:
• Surveillance of antimicrobial resistance can serve as an indicator of the quantitative use of antibiotics in the catchment area covered by the network
• Antimicrobial resistance information, when regularly provided to prescribers, improves the selection process for treatment of community-acquired or nosocomial infections.
• Managers of national or international programs for treatment of infectious diseases, such as acute respiratory infection, diarrheal diseases, sexually transmitted infections, require timely, reliable information related to resistance patterns in causative agents to provide treatment options.
• Regular monitoring of resistance patterns in nosocomial infections is necessary to develop guidelines for the use of prophylactic antibiotics in surgery.
• Information on local or worldwide changes in resistance patterns is necessary in order for health authorities to track epidemics and to make sound recommendations to control outbreaks of infectious disease.6
Many organizations have recognized the importance of surveillance programs to, among other objectives, assist patient diagnosis and treatment, enable infection control in hospitals and communities, and support infection control measures at the regional, national and global levels, to address food safety concerns and to inform drug policy and healthcare decisions.19-21 Surveillance is recognized as the first step to understanding current states and progression of resistance over time. For example, the Danish DANMAP’s surveillance efforts have confirmed that there is an association between the quantities of antibiotics used and the occurrence of resistance in certain bacteria.22 Internationally, surveillance with a broad view of antimicrobial ecology has become recognized as a critical first step to controlling growing resistance.
Analysis of resistance trends from surveillance data has provided considerable evidence in support of the association between antibiotic use and incidence of antimicrobial resistance in human patients.23-25 For example, surveillance of carbapenem-resistant Enterobacteriaceae (CRE) revealed an incidence (particularly in Klebsiella spp.) of 4.6% in hospitals and 17.8% in longterm care facilities for the first half of 2012 (U.S. CDC, 2013). In addition, surveillance programs have identified a significant increase in Streptococcus pneumoniae’s resistance to antimicrobial agents such as penicillins, cephalosporins, macrolides, trimethoprim/sulfamethoxazole, clindamycin, tetracyclines, and chloramphenicol.26-30
Surveillance is … DATA FOR ACTION
… the systematic collection, consolidation, and evaluation of relevant data in order to
…determine patterns of antibiotic consumption
…determine the trends of incidence, abundance, diversity and distribution of antibiotic resistant bacteria and antimicrobial resistance genes.
Monitoring refers to the regular, continuous measurement of i) antibiotic use in human or animal patients and ii) measurement and analyses of specific antibiotic susceptibility in target organisms to discern trends in epidemics of antimicrobial resistance.
Better surveillance is required to both gauge the true scope of the problems and to gain a better understanding of the complex interplay of factors that lead to the development of antimicrobial resistance in human pathogens.
Assessing the patterns of antibiotic use over time establishes trends that may be compared with patterns of resistance. In concert, these data can inform decisions to implement strategies to control the development of antimicrobial resistance. Evidence from several surveillance initiatives demonstrates a correlation between consumption of antibiotics and the development of antimicrobial resistance.31-33 Ideally, bacterial resistance data should be reported using suitable denominators and stratification to ensure that relevant indicators are established.34 Surveillance systems should reliably link diagnosis, pathogens and antibiotic use in order to provide a more informative basis for public health decisions.35 Thoughtful design of surveillance systems reduces bias, improves interoperability (i.e., the ease of integration of one system with another), and promotes usefulness (the ability to take action to protect health based on the information provided by the surveillance system). Consideration of development of performance indicators to monitor the progress of surveillance is also a valuable model.
For the specific question of antimicrobial resistance and utilization surveillance, such systems allow the recognition of worrying resistance trends, recognize connections between utilization and resistance, and identify targets for intervention.
1.3 A Canadian Perspective on Antimicrobial Utilization and Resistance Surveillance
In Canada, there have been a number of reports dealing with aspects of AMR and AMU issued over the past years, as reviewed in Appendix E. The reports range from the 1997 Controlling Antimicrobial Resistance: An Integrated Action Plan for Canadians through to the most recent Canadian Committee on Antimicrobial Resistance (CCAR, now disbanded) reports, including the National Action Plan on AMR in 2004 and the Pan Canadian AMR Consultation Report in 2009. Key recommendations have included:
1. Monitoring of antimicrobial usage in various settings, human and animal.36-39
2. Optimizing appropriateness of antimicrobial usage.40
3. Developing and using standardized formats for surveillance data analysis and dissemination.37
4. Supporting the development of professional competency in antibiotic use.37
5. Developing practice-specific guidelines for prudent use of antimicrobials in humans and animals.36,37
6. Developing a real-time feedback loop to prescribers.41
7. Ensuring that existing and emerging resistance is monitored and that laboratory methodologies are standardized.37 Monitoring of antimicrobial usage in various settings, human and animal.
These past reports have concluded that Canada’s system lags behind other countries in collecting and reporting emerging antimicrobial resistance in Canadian communities, and on data on utilization of these critical medications across human and animal populations.
This current project involved two main data collection methods: a systematic review of published and grey literature on surveillance activities in Canada and internationally, and in-depth interview based surveys of national key experts representing various professional domains. The final component was a review of previous Canadian AMR and AMU reports and publications to inform development of a workable action plan.
It is the aspiration of all participants in this project that this document will serve as a call to action and a road-map for evolving a comprehensive AMR and AMU surveillance program in Canada, starting with integration and collation of existing data as a foundation for the expansion of comprehensive reporting to inform public health actions.
2. Methods
• This project used two data collection modalities—a comprehensive literature review of published and grey literature and an in-depth interview based surveys of national key experts representing various professional domains.
• A systematic search protocol was developed to collect relevant literature gathered from 20 data bases. There were 8931 records identified related to surveillance of antimicrobial resistance and antibiotic utilization, which after screening revealed 129 papers that satisfied most inclusion and quality criteria. Programs described in these papers were analysed.
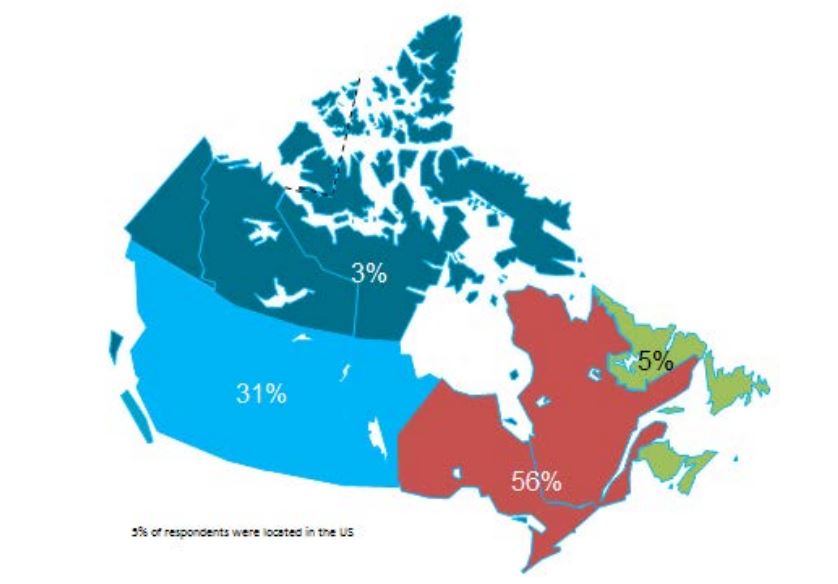
• The survey questionnaire was prepared, pilot tested and issued to 272 experts located in all Canadian provinces and territories. Of these, 146 experts completed the survey questionnaire either in person or by telephone interview.
2.1 Systematic Literature Review
Very few critical evaluations of the major AMR surveillance and antimicrobial use monitoring programs within Canada have been conducted.42 The principal aim of this literature review was to identify and describe the strengths and weaknesses of the major AMR surveillance and antibiotic utilization monitoring programs in Canada, concentrating on antibacterial agents used in human patients and major food animals (poultry, swine and cattle), and to provide a basis for recommendations for improving these programs. A systematic search protocol (Appendix A), designed with a professional medical research librarian, was developed to assemble key information related to AMR and antibiotic use surveillance in Canada and to provide examples from existing programs from around the world for comparative purposes.
Published literature, conference proceedings, and grey literature reports were searched using defined keyword combinations within the limits of specific inclusion and exclusion criteria. Key contacts through academic and professional affiliations provided valuable information regarding research that was not accessible in the published literature. Evidence was gathered to provide answers to the following questions:
1. What exists for surveillance of antibiotic-resistant organisms in Canada, federally, provincially/territorially, and locally?
2. What exists for surveillance of antimicrobial usage in Canada, federally, provincially territorially, regionally, institutionally, and locally?
3. For the above surveillance systems, what information is gathered? To whom is it reported? How quickly is it reported?
4. What international models exist for the collection, reporting and use of the data in monitoring resistance and guiding utilization practice?
5. What provincial/national/international models for legislation and restrictions of use of antimicrobials exist?
6. What surveillance systems for both usage and AMR surveillance have been tried in Canada, what has worked, what has failed and why has it worked/failed?
Twenty databases were searched and a total of 8931 records related to surveillance of antimicrobial resistance and antibiotic utilization were identified. Screening of the search results yielded 580 records for which 335 full-text documents were assessed for eligibility. Two reviewers independently assessed references using a structured form and established process to undertake the quality assessment. Final selected papers were reviewed using the Grading of Recommendations Assessment, Development and Evaluation (GRADE) system43-45 for assessment of quality of evidence. Information collected from a total of 129 documents (Appendix B) met inclusion criteria and data quality objectives and served as the evidence source for the literature review.
2.2 Expert Opinion Survey
A semi-structured survey questionnaire for telephone or in-person interviews was developed to gather information directly from experts who have current knowledge related to surveillance of antibiotic use and antimicrobial resistance in Canada. Following consultation with members of the project steering committee, the initial survey instrument was designed based on answers to some key questions, prepared and pilot tested with anonymous volunteers who were both knowledgeable in the subject matter and in the methodology of conducting qualitative surveys. Five volunteers (including two physicians and one pharmacist) participated in the pilot test phase of the survey preparation that was conducted between October 2012 and December 2012. Some questions were reworded for clarity and redundant questions were eliminated based on comments received from the pilot survey volunteers. The revised survey questionnaire was reviewed and approved for use in the interviews beginning in January 2013 and concluding in May 2013. The survey questionnaire is provided in Appendix C.
Initial key informants were identified from health agencies and institutions across Canada, and included participants in previous conferences, reports and during the systematic search stage of the literature review. They were invited via e-mail to participate in the interviews. By April 2013, respondents from all provinces and territories in Canada had been contacted or interviewed. The survey questions were shared with the participants at the time of receiving their consent and of scheduling the meeting or phone interview. The last question of the survey requested the names of two other experts whose judgments would be valuable for the survey as a ‘snowball’ sampling strategy for enlarging the pool of key informants.
Every interview was transcribed and the data collected were analysed using NVivo 10.0 qualitative analysis software (QSR International Pty Ltd). All survey transcripts were read to determine the overarching themes that emerged. A list of core topics was developed with similar code topics clustered together. The preliminary analysis was conducted by tabulating the frequency at which certain words or phrases were used. As new codes emerged, these were placed in existing theme categories (where appropriate) or a new sub-theme was created. Participants were grouped by discipline (broadly ‘human medicine specialist’ or ‘veterinary medicine specialist’). All findings were compiled and described in narrative form by theme, sub-theme and supporting codes.
3. Description of Canadian and International Surveillance Systems identified in the Literature Review
• Worldwide, there are at least 58 surveillance programs for AMR in human medicine and 21 surveillance programs that track veterinary AMR. Of these surveillance programs, some are/were independently funded projects of finite duration while others are government-sponsored surveillance programs that have been in existence for at least fifteen years.
• There are relatively few surveillance programs monitoring AMU.
• Systematic search of the literature identified 11 Canadian and 58 international ongoing surveillance programs which were evaluated and compared in this report.
• In addition to AMR and AMU surveillance programs, there are several initiatives in Canada and worldwide that are dedicated to knowledge transfer related to the public health threat of antimicrobial resistance.
3.1 Introduction
Many surveillance programs worldwide attempt to map trends in antibiotic use and/or antimicrobial resistance, ranging from small studies of finite duration to large, multi-year ongoing national surveillance initiatives. Our systematic review of 20 databases and grey literature identified 129 papers and reports (out of 8931), with 11 Canadian and 58 international programs described in tabular format in Appendix B. These included surveillance projects of defined duration and some pharmaceutical funded longitudinal resistance studies. Our search captured all programs identified in the expert surveys. More detailed review of a collection of the identified programs is included to provide a good description of existing Canadian initiatives and surveillance models that can supply quality data to health authorities, stimulate cooperation among stakeholders and serve as a platform for exchange of educational information. For readers who want a more exhaustive assessment of international AMR and AMU programs, we direct them to published assessments available.46-49
It is clear there are a number of examples that could serve as models of infrastructure and funding for Canada. Summarized below are examples of ongoing antimicrobial resistance surveillance programs and specific projects from Canada and around the world. Certain programs are highlighted as examples of completeness (DANMAP, NethMap), models for a decentralized model potentially more comparable to the Canadian provincial-federal model (EARS-Net), and examples of innovative funding models (BSAC, CBSN) or because of comparable geography (NARMS). All sustained Canadian projects are included. Other international programs are included here for completeness of the review, but with less detail. For the purposes of this report, “comprehensive” refers to any program that collates resistance and utilization data in a single report or uses a “One Health” model concerning resistance of bacteria across human and animal populations.
Summary tables have been created from data extracted during the literature review and reflect only those data that are publicly available. Where possible for Canadian programs, contact has been made with project leaders to add further information about ongoing and new work that may not be reported. The information from those conversations is included in the text, but not in the summary tables. The criteria for evaluation are based on information extracted from expert interviews (e.g. choice of organisms and antimicrobials) on the components of ideal programs, with support from the literature on the definitions of surveillance. The criteria for evaluation were further vetted by the steering committee.
In Canada, there is one federally funded nation-wide program that looks at both antimicrobial resistance of enteric pathogens in the food chain, and utilization in animals and humans (CIPARS), as well as a regional program that concentrates on human resistance and utilization. In addition, there is a federally funded nation-wide surveillance program for monitoring nosocomially acquired resistant organisms (CNISP) in the setting of overall nosocomial infection surveillance in sentinel hospitals across Canada, which has begun to gather data on utilization, but has not published results to date. Another two programs are collaborations between academic institutions and pharmaceutical industry partners that conduct voluntary prospective sampling from centres located across Canada for AMR surveillance. While some provinces collect data of varying scope in support of surveillance for AMR, to date, only British Columbia provides a formal report of those data. There are provincial and local programs that also provide information locally and initiatives that are being developed for which data are not yet available.
The assessment below addresses specifically the appropriateness of these initiatives as they pertain to comprehensive AMR and AMU surveillance to determine population utilization of and prevalence of resistance to antimicrobials, both overall and in specified microbes and sub-populations of patients. Since these programs may not have been created for that purpose, our assessment is not intended to be a critique of their value for their intended purpose, but rather their contribution to a comprehensive, national AMR and AMU report.
3.2 AMR-AMU Programs in Canada
Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS)
CIPARS collects information on antimicrobial resistance in the three most frequently isolated serovars of Salmonella: S. enteritidis, S. heidelberg, and S. typhimurium (S. newport is sometimes included), from all human clinical isolates through provincial reference laboratories. No data are provided about the provenance of the organisms among human populations. However, given the comprehensive nature of the data (all isolates in Canada), they can be thought to be representative of the population at risk. In addition, data related to the use of antibiotics in human medicine are retrieved via an agreement with IMS Health in Canada (also known as IMS Brogan). IMS Health in Canada is a private company that collects data from retail pharmacies on prescriptions of oral antimicrobials and uses sampling and modelling to predict the overall use of antimicrobials (and other drugs). Data are analyzed by drug and by province. Neither indication nor demographics are included. Data from hospitals are not available, although CIPARS is currently working to gather those data (personal communication).
The CIPARS program also integrates information on the incidence of antimicrobial resistance in food animals into the broader surveillance program by testing susceptibility in enteric pathogens at the on-farm, abattoir and retail levels. At present, there are five veterinary AMR surveillance components within the CIPARS programs: retail meat surveillance (beef, chicken, and pork); abattoir surveillance (beef cattle, chickens, and pigs); farm surveillance (pigs); surveillance of animal clinical isolates (cattle, chickens, pigs, turkeys, and horses); as well as feeds and their ingredients. The target population of consumers of retail meat products in Canada is represented in the retail surveillance component of the program that monitors bacteria of interest and collects information on AMR in the food chain prior to human consumption. The key bacteria of interest in chickens and turkeys are Campylobacter spp., Salmonella spp., Enterococcus spp. (discontinued in 2010), and E. coli while only Salmonella spp. and E. coli spp. are monitored in pork and in beef.
Data on antimicrobial utilization were gathered from the Canadian Animal Health Institute (CAHI), which records 95% of licensed animal pharmaceutical products distributed for sale. Data are aggregated on a class level and cover food, companion and sport animals, as well as fish. However, approximately 30-40% of antimicrobials are used through own-use importation or active pharmaceutical ingredient products which are not recorded. Gross tonnage is reported, but assessment of animal population data is missing. Data on antibiotic use in pigs on CIPARS sentinel farms in the five major swine-producing provinces has been available in CIPARS since 2007; and antibiotic use in broiler chickens is assessed in 2013 data (personal communication).
Quarterly summaries, short reports (containing raw data without interpretation), surveillance bulletins and full annual reports for CIPARS surveillance activities were available on the dedicated website two years after the completion of the data period, however in 2013, it appears that CIPARS reports are now available by request only (i.e. no longer freely accessible on the dedicated website).
British Columbia Centre for Disease Control (BCCDC)
In collaboration with laboratories around the province, the BCCDC produces an annual compilation of resistance trends across a broad array of organisms and antibiotics. These are reported by bug/drug combination and broken down by region/lab. Data are drawn from community and hospital laboratories and reported according to the source of the information. Most B.C. hospital labs provide data to BCAMM, the British Columbia Association of Medical Microbiologists (see Sec 3.3), rather than the BCCDC, so these data are more community focused. There is no explicit mention or analysis of sub-populations (e.g. children, elderly) with respect to antimicrobial resistance, however since data concerning most of the population of the province are captured in a similar manner they are likely to be representative of the population as a whole. In addition, it is likely inherent to the data source (laboratories) that demographics cannot be extracted. BCCDC also provides an annual report on antibiotic utilization in the province using data extracted from the B.C. PharmaNet prescription drug database which tracks all outpatient prescriptions for antibiotics (and other drugs) as they are filled at the pharmacy level. This source limits the assessment of utilization to those drugs that are available orally and excludes those used in hospital. These data are analyzed by sex, age and region and compared to relevant benchmarks. Where possible, utilization with respect to indication is assessed. There are no data specifically on the First Nations population, who may be excluded from community pharmacy data due to the federal responsibility for their medical care. No data are available on veterinary resistance or utilization. Available reports are dated 2012 in which resistance data are current to 2012 and utilization data are current to 2010.
3.3 Programs Reporting Antimicrobial Resistance in Humans in Canada
Canadian Nosocomial Infection Surveillance Program (CNISP)
The Canadian Nosocomial Infection Surveillance Program (CNISP) collects information from 54 sentinel Canadian institutions located in ten provinces. CNISP is designed specifically to assess the rates of acquisition of nosocomial pathogens and nosocomial infections.50-52 Thus, the program provides information on incident infections in hospitalized patients and specifically excludes prevalent and community-acquired infections. Population coverage of CNISP includes all of Canada’s major urban centres and the data compare well to provinces with universal surveillance for nosocomial infections, based on an internal review of representativeness. However, CNISP participants are larger, more urban and more academic than the Canadian average, with over-representation from British Columbia, under-representation from Quebec, and with no representation from the territories (personal communication). Paediatric hospitals are included as well as hospitals that have mixed adult/paediatric population. Elderly and Aboriginal populations are not documented as special populations by CNISP, but should be included (i.e., identifiable) in the general hospital population commensurate with their use of hospitals within large urban centres.
The organisms reported by CNISP are focused on antimicrobial resistant organisms (ARO) of specific concern in nosocomial transmission, including MRSA, VRE, C. difficile and most recently carbapenemase producing organisms (CPOs), as well as specific hospital-acquired infections (not related to specific organisms). Further, because of the nature of the surveillance, limited organism-drug combinations are reported (e.g. S. aureus to methicillin, Enterococcus to vancomycin), instead of more complete susceptibility data that would be necessary for comprehensive hospital surveillance of evolving AMR. CNISP appropriately reports nosocomial infection rates over a denominator of patient days; however it does not report rates of resistance (resistance per number of organisms) or prevalence of resistance (percent of patients carrying a resistant organism). No data are currently available regarding antimicrobial utilization, although data have been gathered from participating hospitals and there is an intention to include these data in future reports (personal communication). Data are current to 2011 on the PHAC website.
FoodNet Canada (formerly C-EnterNet)
This surveillance initiative includes two components: it measures selected pathogens in retail food, agricultural operations and water sources at sentinel sites (one in the Waterloo region of Ontario and one in the Fraser Health Region in British Columbia); and it measures data on cases of food and water borne illness for which there is a legal requirement to report (campylobacteriosis, giardiasis, listeriosis, shigellosis, salmonellosis, and verotoxigenic E. coli). No data are provided about the (human) population provenance of the organisms. However, given the comprehensive nature of the data (all isolates reported to public health labs in Canada), they can be thought to be representative of the population at risk. Specific animal populations at the sentinel sites include: dairy and beef cattle, swine, and broiler chickens. It may be that two sentinel sites are not representative of animal populations across the country. The most recent report available to the public is from 2009.
Canadian Bacterial Surveillance Network (CBSN)
The Canadian Bacterial Surveillance Network (CBSN) performs voluntary prospective collection of isolates from participating clinical laboratories from ten provinces in Canada targeting specific organisms of interest. The dataset includes susceptibility data for Streptococcus pneumoniae and Haemophilus influenzae. Also collected, although not reported on publicly available sources, are resistance of E. coli and Klebsiella species to third-generation cephalosporins, enterobacteriaceae to carbapenems and enterococci to ampicillin (personal communication). Isolates are collected by voluntary submission of all isolates or the first specified number of consecutive isolates of the particular target organism in the study period. Data on patient demographics are not reported in publications accessed, nor is there detail on the nature of the participating laboratories (community, hospital, private, etc.). The latest data available are from 2010.
Canadian Ward Surveillance Study (CANWARD)
Beginning in 2007, the Canadian Ward Surveillance Study (CANWARD) is an ongoing, multi-year program that follows trends of antimicrobial resistance in specific hospitals (10-15 participating hospitals in 8 provinces). Patient information (demographics) is not available in published reports, although the nature of the ward and site of infection is. Isolates representing community populations are only available through isolates submitted in hospital emergency rooms and outpatient clinics.53,54 The program is a pharmaceutically-funded, academic institution partnership. CANWARD provides antimicrobial susceptibility data on the most comprehensive number of organisms (including most from the list of target organisms) and has an interactive website that allows assessment by region of origin. Specific patient populations are not mentioned in reports, although the study is designed to gather a representative sample of the organisms cultured by participating laboratories. Multiple peer reviewed publications have resulted looking at trends in antimicrobial activity.55-68 This program follows the most comprehensive number of organisms of interest. However, the participating institutions may not be representative of the population overall, and a time-interrupted methodology may miss emerging trends. Data are current to 2011.
Toronto Invasive Bacterial Diseases Network (TIBDN)
The Toronto Invasive Bacterial Diseases Network is a population-based surveillance program for selected serious bacterial and viral infections in the Toronto and Peel regions. It was established under contract from the U.S. CDC and includes collaboration between 25 hospitals and 19 microbiology laboratories thought to capture the totality of the region. Current surveillance includes reporting of resistance in invasive isolates of Streptococcus pneumoniae, to specific antibiotics—penicillin, tetracycline, erythromycin, trimethoprim/sulfamethoxazole, amoxicillin, and ceftriaxone. Additional data have included serotypes of Group A Streptococcus, Group B Streptococcus and Neisseria meningitidis. Because invasive (sterile site) isolates are collected, the data may be skewed towards more invasive or pathogenic strains of the bacteria tested. Data regarding population sub-groups is not available; however since this study captures all invasive disease in a geographic area, it can be considered to be representative of that area. Available data are current to 2011 but may have limited distribution.
British Columbia Association of Medical Microbiologists (BCAMM)
The British Columbia Association of Medical Microbiologists (BCAMM) report laboratory data (community and hospital) related to antibiotic resistant organisms—VRE, MRSA, S. pneumoniae and ESBLs—throughout the province. These data encompass a defined set of nosocomially important specific bacteria-antimicrobial combinations, similar to the CNISP program, but rates of resistance (resistant isolates/total isolates) are available. Reporting has been conducted annually since 2002 and information is made available to the Provincial Health Officer, the BCCDC and other groups interested in surveillance of antimicrobial resistance in the specified pathogens. These data are included in the BCCDC report on antimicrobial resistance as well as in a free-standing report. The report and its content are a voluntary production of a professional organization, done without funding. The most recent available report provides data from 2011.
3.4 International Comprehensive AMR-AMU Programs
Comprehensive surveillance systems of AMR and AMU are generally viewed as the ideal state (see experts survey below). These programs collect data on human and animal AMR and AMU, and many adopt the One Health approach of including veterinary and human populations in reports. These initiatives are most developed in Europe, although initial attempts to develop aspects of such programs have extended to North America (CIPARS in Canada and NARMS in the U.S.A.). Some developing nations (e.g. India, Columbia) have begun to initiate such surveillance programs as well.
DANMAP
DANMAP has adopted the ‘farm to fork’ approach for conducting surveillance activities which allows antimicrobial resistance in zoonotic and indicator species (organisms that are ubiquitous in animals, food and humans) of bacteria to be tracked along the food chain and monitors antibiotic consumption in animals and humans.69 It is a collaboration between the Danish Ministry of Food, Agriculture and Fisheries and the Danish Ministry of Health which produces an annual report that examines Antimicrobial resistance in human and animal pathogens, zoonotic bacteria and indicator species.70
Bacterial isolates are collected from healthy animals at the time of slaughter in addition to diagnostic submissions and some subclinical cases. The DANMAP program includes surveillance of pet animals (dogs, cats, birds, mice and guinea pigs) and horses, in addition to food-producing animal species (poultry, cattle and swine). Bacteria from food samples (both produced in Denmark and imported) are regularly collected by the regional veterinary and food control authorities.
Clinical bacteria in humans are captured from microbiology laboratories from 12 out of 13 departments, representing 95% of the population of Denmark. Organisms isolated in blood, urine, and CSF samples (depending on the organism) from humans that are monitored for antimicrobial resistance include Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Streptococcus, Enterococci, Bacteroides fragilis and Staphylococcus aureus. In addition, ESBL-producing bacteria, Neisseria gonorrhoeae and Clostridium difficile have been included in the surveillance program since 2009. Antimicrobial resistance monitoring in zoonotic bacteria focuses on Salmonella spp., C. difficile and Campylobacter spp. in both human and animal hosts, including an analysis of the likely species origin of human disease. Indicator species monitoring includes sampling for resistance patterns of E. faecium, E. faecalis, and E. coli in animal fecal samples and retail meats.
The DANMAP surveillance program collects data on antimicrobial agents registered for use to treat bacterial infections in humans from all five health care regions in Denmark. The Danish Medicines Agency (the Lægemiddelstyrelsen, similar to the U.S. Food and Drug Administration) has monitored antibiotic drugs prescribed to individual patients since the early 1990s. Over time, DANMAP has reported usage data together with antimicrobial resistance data.. This has allowed specific problems connected with antimicrobial consumption on human therapy to be identified.69 Data are collected from primary care settings and hospitals and reported in terms of defined daily doses (DDDs) as the primary unit of measure. Consumption data for primary care and for hospitals are reported as DDD per 1,000 inhabitants per day (DID), DDD per 100 occupied bed-days (DBD) and DDD per 100 admissions (DAD). Usage data are converted to kilograms of active ingredients of antimicrobial agent to allow comparison between consumption of antimicrobials in human and in animal patient populations.
Given the breadth of coverage of the country by surveillance (>95% of the population), these data are representative. However, in the publicly available reports there is no breakdown by demographic group or sub-population, and so, for example, results for Denmark’s Inuit (i.e., Greenlanders), cannot be compared directly to Canadian Inuit populations.
Data related to antimicrobial use in animals have been reported since 2001.71 In Denmark, prescriptions are required for all therapeutic medicines and VetStat, a national registry, collects the data for all medicines prescribed by veterinarians for use in animals including the Nordic Item Number, veterinarian identity, amount, date of sale, farm or practice identity, species, age group and disease group.72 Consumption is measured in grams of active ingredient or in number of doses and this value describes the numerator in consumption rate equations. The doses are species-specific and described in terms of “Defined Animal Daily Doses” (ADD). For overall consumption comparisons, kilograms of active ingredient are reported for veterinary antibiotics. The denominator that is used in DANMAP is described in terms of animal production, either by kilograms of meat produced or number of animals produced.73 The denominator used for comparing selection pressure between species describes the population at risk (biomass-year-at risk). This value accounts for differences in both lifespan and in body mass and is similar to the denominator used in human pharmaco-epidemiology (inhabitant days, DID) with the exception of the time interval (year vs. 1000 days).72
DANMAP has produced annual reports summarizing results and program activities every year since 1996. Many reports of individual studies have been published over the years and research associated with the DANMAP surveillance system has made significant contributions to the peer-reviewed literature.73-78 The latest available data are from 2012.
The European Centre for Disease Prevention and Control
The ECDC runs four projects to create a human AMR and AMU surveillance program:
- EARS-Net (formerly The European Antimicrobial Resistance Surveillance System) collects and collates data on isolates from human blood and cerebral spinal fluid cultures including Streptococcus pneumoniae, Staphylococcus aureus, Enterococcus faecalis, Enterococcus faecium, Escherichia coli, Klebsiella pneumoniae, Acinetobacter and Pseudomonas aeruginosa from 28 member countries;
- The European Surveillance of Antibiotic Consumption (ESAC) monitors antibiotic consumption in both ambulatory and hospital care in 24 (and two non-member states) member countries of the European union.79
- The Centre européen d’études pour la santé animale (CEESA), in collaboration with EARS-Net, conducts pharmaceutical industry sponsored veterinary surveillance of antimicrobial resistance in food animals.
- The European Surveillance of Veterinary Antimicrobial Consumption (ESVAC) gathers data on antimicrobial consumption in animals.
The European Antimicrobial Resistance Surveillance System (EARSS) was transferred to the European Centre for Disease Prevention and Control in January 2010 and renamed EARS-Net. This program is a part of The European Surveillance System (TESSy), and is supported by European Union legislation (Decision No. 2119/98/EC). Annual reports including trend analyses are published by the ECDC and are publicly available on the ECDC website. There is significant heterogeneity in terms of the proportion and make-up of the patient population covered in each member country and over time. Because of annual differences in coverage within the same country, sensitivity analyses are performed to test the robustness of trend data. Because of the invasive nature of the specimens, the data may be skewed towards more invasive or pathogenic strains of the bacteria tested. Further use of data is possible as the metadata set of all variables that are reported through the TESSy system are available for the use of European Union member states. The most recently published report is from 2012.
ESAC data are collected from national sales and/or reimbursement data for medications given systemically (excludes topical agents). Data provided by different countries differ according to the ability of individual countries to collect data, however almost all countries report population coverage close to 100%.80 Data are collated centrally and calculated as defined daily doses (DDDs) per 1,000 inhabitantdays for community and hospital prescriptions. Coverage of specific antibiotics is not as comprehensive, nor is it as finely focused as individual country reports. However, this likely represents the geographically large area covered by the report, and the differences in consumption between countries. ESAC also reports on the consumption of antimycotic and antiviral agents. There are no data linking consumption to indication, except for antiviral drugs, nor on specific patient populations. Veterinary populations are not included in this report.
Surveillance of antimicrobial resistance in animals by the ECDC combines four programs: VetPath (follows antimicrobial susceptibility of major bacterial pathogens in food animals); European Antimicrobial Susceptibility Surveillance in Animals (EASSA) (monitoring susceptibility in food-borne and commensal bacteria in food animals); ComPath (examining antimicrobial susceptibility in major bacterial pathogens in companion animals); and MycoPath (evaluating antimicrobial susceptibility of major disease-causing mycoplasma species from food animals).49 Isolates are collected from healthy and diseased animals originating in several participating countries in Europe. Species studied are Salmonella spp., Campylobacter spp., E. coli and Enterococci.
ESVAC collects data from nine European nations through multiple mechanisms including wholesalers, pharmacies, veterinarians and marketing authorization holders. Species covered include pigs, poultry, cattle, sheep, goats and horses and are expressed as milligrams per population.
The most recent reports for ESAC and EARS-Net provide data from 2010, while reports from ESVAC provide data to 2009.
NethMap and MARAN
The Netherlands now has a comprehensive single report including data on human and animal resistance since 2012, combining two programs, NethMap and MARAN.
NethMap is the program name for a cooperative effort between members of SWAB (Stichting Werkgroep Antibioticabeleid) and the Centre for Infectious disease control (Cib) at the National Institute for Public Health and the Environment (RIVM) that conducts surveillance on antibiotic use and antimicrobial resistance in common human pathogens isolated in the Netherlands. Patterns of antimicrobial resistance are reported for Escherichia coli, Klebsiella spp., Enterobacter spp. Proteus mirabilis and Pseudomonas aeruginosa, staphylococci, enterococci and respiratory pathogens; occurring either in particular patient populations (e.g. ICU) or infection types (e.g. urinary tract). Patient populations that are monitored include hospitalized patients, patients visiting general practitioners, patients in nursing homes and outpatient departments. Data are separated by demographics (i.e. age) and type of care (e.g. community, long-term care, intensive care units, etc.) however children are not a subpopulation that is specifically targeted.
The surveillance program that monitors antimicrobial resistance and antibiotic usage in food animals in the Netherlands is named MARAN. Salmonella spp. and Campylobacter spp. are the principle food-borne pathogens measured in poultry and in swine. Broilers (chickens), eggs, dairy cattle, milk, veal calves, pork and turkeys are monitored for the selected pathogens with testing conducted at the reference laboratory. E. coli and enterococci are monitored for resistance as indicator organisms for the commensal gut flora.81 Listeria monocytogenes has been included in the surveillance program since 2004. In cattle, E. coli, Staphylococcus aureus, Klebsiella spp. and Enterobacter are monitored for resistance. Resistance in isolates of Salmonella spp. collected from animal feed, turkeys, horses, ducks, pigeons and reptiles are also reported. In addition to Campylobacter spp. and Salmonella spp., Shiga-toxin producing E. coli and ESBL isolates are monitored in cattle and in human isolates.82 E. faecalis and E. faecium isolates are now included in the indicators of commensal gut flora and these are monitored in raw meat products, vegetables, fruits and herbs. The MARAN surveillance program also collects resistance data for important veterinary pathogens including bovine mastitis pathogens E. coli, coliform bacteria (Enterobacter, Klebsiella and other species), Staphylococcus aureus, Brachyspira hyodysenteriae and Mycoplasma synoviae.
Since 2004, the Netherlands has collected data through a continuous monitoring program at the farm level that is managed by the Agricultural Economics Research Institute (LEI) for the usage of antibiotics at dairy, pig and broiler farms. Sales data from the pharmaceutical companies offer a general estimate of the veterinary usage of antibiotics with more detailed information provided from the ‘Farm Accountancy Data Network.’ This data network provides information related to exposed animal populations, specific species, characteristics of the farms and total number of animals. Veterinary medicines incorporated into animal feed are included in this sales data.
Rather than express the exposure data of veterinary drugs as kilograms of active substance (the numerator), the unit of measurement is the number of daily doses. This is calculated as the quantity of the veterinary medicinal product divided by the approved dose of the given medicine.81 The denominator describes the population at risk and the period of time during which consumption is measured. This is calculated as total number of animals at risk of being exposed to antibiotic per year.
The total number of kilograms of antibiotics (as active ingredient) sold in the Netherlands at the level of pharmaco-therapeutic group is reported by the Federation of the Dutch Veterinary Pharmaceutical Industry (FIDIN). The usage data are based on the sales data of FIDIN members. Prior to 2009, sales data provided information for total sales of active ingredient for all animals. However, since the implementation of the European Surveillance of Veterinary Antimicrobial Consumption (ESVAC) data collection protocol, the levels of active antibiotic ingredients take into account salt and ester formulations as well and all calculations are expressed in international units.82 More detailed information about usage patterns on a per-animal species basis is also available.
The most recent report available is for 2012 (as a joint report between NethMap and MARAN).
National Antimicrobial Resistance Monitoring System (NARMS)
NARMS is a collaboration between the U.S. Food and Drug Administration (FDA), the Foodborne diseases active surveillance Network (FoodNet) of the CDC and the U.S. Department of Agriculture (USDA). The program collects specimens of Salmonella from all 50 state laboratories and Campylobacter, derived from a representative sample of human clinical cases from participating state laboratories (representing approximately 15% of the population). Human samples of E. coli 0157 and Vibrio samples are also gathered through the FoodNet program and reported with NARMS data.
Since 1997, the animal component of the program has collected data from animal isolates of Salmonella as the sentinel organism with other key pathogens included in subsequent years (Campylobacter (1998), E. coli (2000), and Enterococcus (2003)). The animal isolates are gathered from federally inspected slaughter and processing facilities and USDA National Animal Health Monitoring studies on farms located across the country. Diagnostic animal specimens, food-producing animals at time of slaughter and healthy on-farm animals are the sources of the isolates monitored in the NARMS program.
Since 2002, monitoring retail meat has been a component of the NARMS program through cooperation between the CVN, CDC and FoodNet laboratories. Some selected sites also culture retail meat samples for E. Coli and Enterococcus. NARMS data are available for the monitoring of animal isolates as annual reports summarizing data collected from chickens, turkeys, cattle and swine (since 2005); summary tables and reports for individual bacterial organism categorized by major animal source, clinical status and years; and interactive data query pages. Annual reports are available on-line since 1996. Demographics are not available for human isolates, but based on random sampling methodology is likely to be representative of the population in the participating states. Data are not available on antimicrobial consumption in the NARMS report; however a complementary FDA report provides gross tonnage of antibiotics purchased for use in food-producing animals. Linkages to use in specific animals species are not provided. Component reports are current to 2011.
Norwegian Surveillance System for Antimicrobial Drug Resistance (NORM) and NORM-Vet
This surveillance program has been collecting data since 2000 and is managed by the Norwegian Institute for Public Health. Isolates are tested through the Infection Control program at the University Hospital of North Norway and reported through NORM. In addition to antimicrobial resistance data, the NORM program collects information related to antibiotic use in hospitals and long-term care facilities. This surveillance system provides the data for antibiotic use in Norway and reports data related to resistance in key pathogens and nosocomial infections to the ESAC. The program has participated in the European EARS-Net since 2004.
NORM-Vet data are publicized in the same report, and include data on indicator bacteria in animals and food (E. coli from meat, MRSA from bovine mastitis and swine, and ESBL from swine and wild reindeer.) AMU data includes sales of antimicrobial veterinary medicinal products for therapeutic use in food animal production, companion animals and farmed fish. Cocciodostat feed additives are reported as well. No antimicrobial growth promoters have been used in Norway since the food animal production industry abandoned their use voluntarily in 1997.
Swedish Strategic Programme against Antibiotic Resistance (STRAMA) and Swedish Veterinary Antibiotic Resistance Monitoring (SVARM)
SWEDRES began as a joint venture of STRAMA and SVARM, providing joint reports of antibiotic utilization and resistance in humans and resistance in animals.
Data are gathered on antimicrobial consumption, reported by age stratum and geographic region with some data on indication for specific infections (e.g. respiratory). Resistance data are reported for specific bug-drug combinations (e.g. S. aureus), and more comprehensively for organisms of interest (e.g. E. coli in urinary tract infections). In addition to surveillance, STRAMA is a model which combines surveillance, stewardship, feed-back of own data and performance, public benchmarking against “STRAMA indicators”, point-prevalence and diagnose-prescribing surveys, and compliance to national treatment guidelines. STRAMA has conducted several large-scale diagnosis-prescription surveys in hospitals, in primary care settings and in nursing homes.83 The pharmacy monopoly in Sweden has enabled the collection of a well-defined data set on the sales of antibiotics throughout the country, although inadequate IT systems have, in some cases, impaired the surveillance system.84 SVARM collects data on antimicrobial usage in animals from overall sales data. The two programs provide a combined report since 2012, and that is the date of the most recent reported data.
Finnish Study Group for Antimicrobial Resistance (FiRe Network) and Finnish Veterinary Resistance Monitoring and Consumption of Antimicrobial Agents (FINRES-Vet)
The Finnish Study Group for Antimicrobial Resistance has operated as a nationwide network since 1991 to monitor antimicrobial resistance in clinically important human pathogens and antimicrobial consumption. The program tracks resistance in isolates of several important human pathogens derived from urinary tract, respiratory tract, soft tissue and blood borne infections of hospitalized patients and outpatients.85-89 Data on the consumption of antimicrobials has been collected by the National Agency for Medicines and the statistics were usually derived from sales data from wholesalers to pharmacies. The data represent the annual consumption levels from each central hospital district expressed in defined daily dose (DDD) per 1000 inhabitants, per day. At the community level, the consumption of antibiotics for outpatient care in some central hospital districts in Finland was also monitored. In addition, Finland has a data network to collect data on veterinary resistance and antimicrobial consumption which provides annual reports, the most recent of which is from 2009. Data from the human program appear to be reported in peer-reviewed publications rather than in regular reports.
COIPARS: Columbian Integrated Program for Antimicrobial Resistance Surveillance
This developing program has so far published on Salmonella in retail chicken and broiler farms in Columbia, in 2010 and 2011, and is operating under a One Health approach.
3.5 International Programs that Report Antimicrobial Resistance in Humans
Surveillance programs that monitor antimicrobial resistance in pathogens have been conducted for many years in the United States, European countries, Russia, Japan, Australia, New Zealand, South Africa and some Asian countries. The World Health Organization has been responsible for surveillance efforts undertaken in developing nations although in many cases, national surveillance programs for individual countries do not exist. Moreover, in several regions of the world antibiotics can be sold without prescription or oversight by any health-care professional thereby making efforts to reduce the risk to public health of development of antimicrobial resistance particularly challenging.90 A recent initiative in India, The Chennai Declaration, has started to address the issue of over-the-counter antimicrobials and resistance.
The WHONET software was developed by the WHO Collaborating Centre for Surveillance of Antimicrobial Resistance (Harvard University) for the management and analyses of antimicrobial susceptibility laboratory data. The database software has been implemented in local and national surveillance programs of more than 90 countries thus far.
British Society for Antimicrobial Chemotherapy Resistance Surveillance Project (BSAC)
Surveillance of resistance in the United Kingdom and Ireland has been conducted by the British Society for Antimicrobial Chemotherapy Resistance Surveillance project, which employs a ‘consortia-style’ funding model in cooperation between pharmaceutical companies and the BSAC. The program monitors antimicrobial resistance in community-acquired respiratory tract and bloodstream infections.91 Two distinct programs are part of the surveillance project: the Bacteraemia program gathers 7–14 consecutive samples of the most commonly isolated organisms (S. aureus, CNS, S. pneumoniae, Streptococci, Enterococci, E. coli, Klebsiella spp., Proteas spp., Pseudomonas spp., Enterobacter spp. and Serratia spp.) from 40 representative centres. The Respiratory program collects isolates of communityacquired pathogens (S. pneumoniae, H. influenzae, M. catarrhalis) and samples of isolates of hospital-acquired lower respiratory infections (S. aureus, Pseudomonas spp., Acinetobacter spp., enterobacteriaceae) from the same centres during a limited time period annually. Samples are tested in a centralized laboratory for antimicrobial susceptibility. Demographics including ward or outpatient clinic, site of infection, and patient demographics are collected. There are no formal reports; rather information is transmitted in an interactive antibiogram on the website, which has data to 2012, and through peer-reviewed publications. The most recent publications for bacteremia data are from 2011 and for elements of respiratory pathogen data are available from 2012.
Active Bacterial Core (ABC) Surveillance System, CDC Division of Bacterial and Mycotic Diseases
This program determines the incidence, epidemiological characteristics, and microbiology of invasive disease due to Haemophilus influenzae, Neisseria meningitidis, group A Streptococcus, group B Streptococcus, Streptococcus pneumoniae and methicillin-resistant Staphylococcus aureus in numerous large diverse U.S. populations, which represent a population of about 42 million. All laboratories in the geographic area participate and there is central laboratory confirmation, to minimize surveillance bias. Population census data are used as denominator data, which is a fairly unique characteristic of this system. Data are provided by yearly reports of each pathogen (with approximately 18 months lag time) with age, race, geography, and clinical syndrome data, and in scientific publications. There are ongoing special projects for pathogens of particular interest.
Greek System for the Surveillance of Antimicrobial Resistance
The rates of antimicrobial drug resistance in Greece are among the highest reported in Europe.92 In Greece, the National Electronic System for the Surveillance of Antimicrobial Resistance has collected data primarily from hospitals but also outpatient settings since introduction of the program in 1997.93 The WHONET software has been used to process the data and information resulting from the surveillance efforts in Greek hospitals for resistance of several key pathogens to antibiotics including ampicillinsulbactam, piperacillin-tazobactam, ceftazidime, cefepime, imipenem, amikacin and ciprofloxacin.94 Carbapenem-resistant Gram-negative species, pan-resistant Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa are now reported,95-97 as well as the on-going monitoring of MRSA and VRE in hospitalized patients and extended care facilities.
3.6 Surveillance of Antimicrobial Resistance in Veterinary Medicine
Human and veterinary medicine have the shared responsibility for preserving the efficacy of antibiotics used for treatment of infections and preventing the spread of antimicrobial resistant organisms. Animals and humans can act as hosts to the same species of bacteria and thus the same classes of antibiotics, and in some cases the same drugs, are used for similar therapeutic purposes. Furthermore, farm animals represent a model system where the potential of candidate policies for reduction of antimicrobial usage can be evaluated at the population level, with further relevance to policies in humans.98 Early discussions focused on surveillance of antimicrobial resistance in bacteria of animal origin in Europe proposed three levels: i) veterinary pathogens that are considered to be bacteria under greater selection pressure; ii) bacteria of the intestinal flora that are considered to be a reservoir of bacteria under the indirect selection pressure exerted by exposure to antibiotics; and iii) the zoonotic bacteria that can be transmitted to humans via direct contact or consumption of contaminated food products.99
Review of national surveillance programs have recommended harmonization among the various programs with respect to testing methods that are used, bacteria species that are monitored and the antibiotics that are tested.100
Leading health organizations and agri-food businesses agree that national and international surveillance systems for veterinary antibiotic use and antimicrobial resistance in animals must be developed and/or improved to preserve human food safety. Recommendations have been made to incorporate surveillance of farmed aquaculture species into existing Canadian pathogen-based surveillance programs at modest cost in order to ensure seafood safety.101
A ranking system, originally introduced by the Australian Joint Expert Committee on Antibiotic Resistance102 was developed to describe the range of human health consequences of exposure to antibiotic drugs used in veterinary treatment of food animals. This ranking system formed the basis for categorization of antibiotic drugs developed by the Veterinary Drug Directorate of Health Canada (Appendix F). The category I drugs listed in Appendix F are those that are most important for treatment of life-threatening infections in human medicine while category IV drugs are not used in human medicine.
3.7 Veterinary Antimicrobial Resistance Monitoring Programs in Canada
Ministère de l’Agriculture, des Pêcheries et de l’Alimentation du Québec (MAPAQ)
Beginning in 1993, the Ministry of Agriculture, Fisheries and Food of Quebec (MAPAQ) has collaborated with the Faculty of Veterinary Medicine at the University of Montréal to conduct surveillance of antimicrobial resistance in food animals. Passive surveillance was undertaken between 1993 and 1998 to evaluate trends of antimicrobial resistance in food-borne pathogens of importance to public health including Salmonella spp., Escherichia coli and Staphylococcus spp. in poultry, cattle and swine. A number of studies have been conducted through MAPAQ and since 2004 surveillance data of Salmonella spp. in food animal products and retail meats in Quebec have been contributed to the CIPARS program. On-going active surveillance activities in the province of Quebec are conducted by MAPAQ and the Faculty of Veterinary Medicine at the University of Montréal, with reports summarizing antibiograms, resistance patterns, and resistance profiles in important zoonotic and veterinary pathogens for poultry, swine and cattle now produced on an annual basis.103
Université de Montréal – Faculté de médecine vétérinaire
Among the research conducted through the Faculty of Veterinary Medicine at the University of Montreal are projects that investigate the incidence and transmission of antimicrobial resistance in animals. Some of these studies are funded by NSERC and involve active surveillance of antimicrobial resistance in poultry, swine and small companion animals. Few current studies involve passive surveillance and only in some cases is information related to the use of specific antibiotic drugs included in the projects. Several studies are conducted in collaboration with MAPAQ and include surveillance of MRSA, Enterococcus spp. and Clostridium perfringens.
University of Guelph – Ontario Veterinary College
The Centre for Public Health and Zoonosis (CPHAZ) is part of the Ontario Veterinary College located at the University of Guelph. Antimicrobial resistance is one of the six core thematic areas of study at CPHAZ where Canada’s largest capability in AMR research at the human-animal-environment interface has operated for many years. Some research projects conducted at the Ontario Veterinary College include active surveillance of antimicrobial resistance in livestock, companion and sport animals and some exotic animal species. CPHAZ hosts an extensive on-line database of publications focused on antimicrobial resistance in animals with specific information on risk assessment, diagnostic methods, prevalence of zoonotic infections, molecular epidemiology, humans associated with animals, antimicrobial drug use and therapy, and antimicrobial guidelines and policy.
B.C. Ministry of Agriculture
The British Columbia Ministry of Agriculture (formerly the Ministry of Agriculture, Food and Fisheries) collects data for the surveillance of antimicrobial resistance, primarily for isolates from poultry and fish aquaculture. Some monitoring activities provide information for the CIPARS and FoodNet Canada programs as well. BCMAFF is also engaged in antimicrobial stewardship activities that target the agricultural and veterinary communities. Some data are publicly available and some provincial surveillance data is available upon request.
3.8 International Veterinary Antimicrobial Resistance Monitoring Programs
National surveillance programs for monitoring antimicrobial resistance in veterinary medicine have been in existence for several years in Europe (including Austria, Belgium, Denmark, Finland, France, Germany, Ireland, Italy, the Netherlands, Portugal, Spain, Sweden, and the United Kingdom), the United States, Canada and Japan. However, there is a lack of harmonization between many of these programs which makes global comparison of data and resistance trends among countries problematic.104 Monitoring programs can vary considerably between nations based on differences in agricultural practices, monitoring needs and antimicrobial uses and guidelines.105 It has been observed that antimicrobial surveillance systems for monitoring resistance in veterinary medicine in some European nations are not harmonized with respect to methodology, interpretive criteria, epidemiological cut-off values or even an agreed upon definition of resistance.106 Some surveillance programs do have commonalities. For instance, the same antimicrobial agents are monitored for Salmonella (i.e. gentamicin, chloramphenicol,34 nalidixic acid and oxytetracycline) in DANMAP, JVARM, NARMS, MARAN and CIPARS.107
GERM Vet/BfT-GermVet monitoring program
The German national monitoring program (GERM Vet) is conducted by the Federal Office of Consumer Protection and Food Safety (Bundesamt für Verbraucherschutz und Lebensmittelsicherheit, or BVL) in Berlin. GERM Vet has been operating since 2001, and concentrates its surveillance efforts on target bacterial pathogens associated with infectious disease conditions in food-producing animals. GERM Vet is complemented by the BfT-GermVet monitoring program, under the direction of the Federation for Animal Health (Bundesverband für Tiergesundheit, or BfT). The BfT-GermVet monitoring program has been collecting susceptibility data of bacterial pathogens from companion animals since 2003.108 It tests isolates collected from horses, dogs, and cats, and bacteria from diseased pigs and cattle that have not already been tested in the GERM-Vet program for susceptibility to several antimicrobial agents and combinations of antimicrobials.109 Specific resistance genes are also measured in isolates collected from food animals and companion animals in these surveillance programs.
Italian Veterinary Antimicrobial Resistance Monitoring (ITAVARM)
Like most other European Union member countries, Italy began its program for monitoring antimicrobial resistance in 2001 following the commencement of the IV EC Framework Program on Antibiotic Resistance in Bacteria of Animal Origin (ARBAO). Prior to this time, there was little standardization among testing methods and antibiotics that were monitored and few quality control measures were used to ensure the precision and accuracy of the data. Since 2001, considerable improvements have been made in veterinary antimicrobial resistance surveillance in Italy in these aspects of testing and data collection. Italy is among the European Union member nations who participate in the European Union-wide ARBAO surveillance initiative. The Veterinary Reference Centre for Antibiotic Resistance, in collaboration with the Istituti Zooprofilattici Sperimentale, monitor food animals (cattle, swine and poultry) and companion animals (dogs, cats, horses) for three categories of bacteria from isolates collected from animals throughout all regions of Italy.109 The three bacterial categories are: animal pathogens (Pasteurellaceae, coagulase-positive staphylococci, Streptococci, Escherichia coli); zoonotic bacteria (Salmonella spp., E. coli); and indicator bacteria (Enterococcus spp., E. coli). Information related to the veterinary use of antibiotics is not collected through this program.
Japanese Veterinary Antimicrobial Resistance Monitoring System (JVARM)
Established in 1999, the Japanese Veterinary Antimicrobial Resistance Monitoring System (JVARM) collects data to study trends in resistance and the association between use of antimicrobial agents and resistance and for risk assessment and management. The JVARM program tracks bacteria primarily categorized as animal pathogens (generally derived from diseased animals) and zoonotic and commensal bacteria (collected from apparently healthy animals). Key bacteria species that are monitored in this longitudinal program include Salmonella, Campylobacter, E. coli, Enterococci and some animal pathogens.49,110,111 The zoonotic and indicator bacteria are isolated from faecal samples collected from cattle, pigs, broilers and layers. Six samples per animals are collected in each prefecture every year. In addition, the JVARM program collects information related to the quantities of veterinary antibiotics that are used, based on calculations from sales records from pharmaceutical companies that they are required to submit to the National Veterinary Assay Laboratory (NVAL) under the Pharmaceutical Affairs Law.112 This information includes data on the annual sales volume (weight of active compound) by substance, class, and animal species. Data from JVARM reports have demonstrated a relationship between therapeutic antimicrobial use and the incidence of antimicrobial resistance in bacteria collected at the farm level.113,114 The Ministry of Agriculture, Forestry and Fisheries (MAFF) in Japan is responsible for animal husbandry but not food hygiene, and thus bacterial isolates are collected from food animals at the farm level but not in food products
Conversations related to the use of antibiotics in fish are often focused on treatment of infections in finfish species raised through aquaculture. However, antibiotics are frequently used for therapeutic treatment of ornamental fish species kept as pets and several common drugs such as tetracyclines, amoxicillin, sulfamethoxazole / trimethoprim and ciprofloxacin are available over-the-counter at many large pet supply chains. In fact, some internet blogs and related fora (e.g. preparednesspro.com or outdoorsdirectory.com) list pet supply businesses that sell antibiotics intended for treatment of aquarium fish, and discuss the potential for human use of these products, describing them as the same as what physicians prescribe, but cheaper.
There are 195 Livestock Hygiene Services Centres which report to prefecture offices throughout the country that participate in the JVARM program. The NVAL serves as the national reference laboratory, is responsible for compiling and analysing data to be reported to MAFF and conducts research into mechanisms of antimicrobial resistance and molecular epidemiology. Standardized techniques are used for the isolation, identification and susceptibility testing of the target bacteria.
Other veterinary antimicrobial resistance surveillance programs include:
• French Antimicrobial Resistance Monitoring in Bacteria of Animal Origin (Farm)
• Veterinary Monitoring of Antimicrobial Resistance in Spain (VAV)
• U.K. Animal Health and Veterinary Laboratories Agency (AHVLA)
• Belgium Veterinary and Agrochemical Research Centre
• National Reference Laboratory (Ireland) (Antimicrobial Resistance, Food, Feed & Health)
• Global Foodborne Infection Network (formerly Global Salm-Surv)
3.9 Surveillance of Antibiotic Utilization in Veterinary Medicine in Canada
Veterinarians are responsible for ensuring the well-being of animals that are under their care while protecting the health of human and animal populations. In doing so, veterinarians have an obligation to promote strategies in disease prevention, to encourage prudent use of antimicrobials, and to communicate the potential consequences of antimicrobial therapy (including the development of AMR) to animal owners and managers.112 Reliable data for antimicrobial utilization in animals are not publicly available, and thus it is difficult to determine the actual quantities of the various antibiotic drugs and the purpose for which they were used.115,116 Some studies have been conducted through existing surveillance programs worldwide to evaluate veterinary antimicrobial use, usually by reviewing sales data that is reported in various countries.
Recommendations have been made that ideally, usage data should be collected for each animal species and for each product category while taking into account the dosage for each antimicrobial used and the duration of treatment.117
Currently in the United States, there is no comprehensive surveillance system for gathering data on antimicrobial use in animals with the exception of limited data collected by the National Animal Health Monitoring System (NAHMS) and the U.S. Department of Agriculture.118
Canada
There are no nationwide surveillance programs for collecting data related to the utilization of antibiotics in veterinary medicine in Canada. Antibiotics that can be added to livestock feed under Canadian regulations are listed in the Compendium of Medicating Feed Ingredients Brochures (CMIB). The lists are specific to each animal species to treat specific conditions and include the required withdrawal times for safe drug clearance after treatment is completed. Currently, antibiotics can be purchased directly by animal owners and food animal producers for their own use without prescription by a veterinarian. Therefore, it is not possible to accurately estimate the amount of antibiotics that are purchased for own use and there are no programs or standards currently in place to assure quality of the purchased products. For several years in Canada, recommendations have been made that this ‘own-use loophole’ should be closed.
CIPARS has obtained preliminary information on the crude mass of antibiotics used in some sectors of Canadian agriculture. In addition, some information related to the use of antibiotics in food animals in Quebec is available through MAPAQ.
British Columbia Ministry of Agriculture
The Ministry of Agriculture in British Columbia monitors use of antibiotic drugs in poultry and in salmon aquaculture. Of note in the province of British Columbia, the salmon aquaculture industry has voluntarily adopted the practice of administering antibiotics with prescription from veterinarians since 2006.
The University of Guelph – Ontario Veterinary College
Some studies conducted through the Ontario Veterinary College at the University of Guelph have collected data related to the use of antimicrobials in animals. Most of the utilization data are linked to projects that are finite in duration (one to four years). Recent reports include investigations of antibiotic use in dogs and cats in Ontario,119 sheep in Ontario,120 and swine in Alberta.121
3.10 Knowledge Translation Programs
CANADA
National Collaborating Centre for Infectious Diseases (NCCID)
The National Collaborating Centre for Infectious Disease (NCCID) is one of six centres that are funded by the Public Health Agency of Canada. The NCCID has a mandate to foster linkages between health professionals engaged in activities related to the protection of the Canadian population from infectious disease. The NCCID facilitates knowledge translation and research on infectious disease to provide the basis for sound evidence-based decision making in public health programs and the policy arena.
Comprehensive Antibiotic Resistance Database (CARD)
Hosted by the Michael G. DeGroote Institute for Infectious Disease Research at McMaster University, the Comprehensive Antibiotic Resistance Database (CARD) is a bioinformatics database of resistance genes, their products and associated phenotypes. As part of a collaborative project between the United Kingdom and Canada, the database is intended to provide a unified source of information about resistance genes including molecular, clinical and surveillance data. At the genome and plasmid level, CARD has focused on MRSA and Acinetobacter baumannii as well as a few examples of other bacterial species. The database was developed as a tool to study the genetics and genomics of antimicrobial resistance and how it affects bacterial populations, ecology and clinical therapy.122
gFARAD
The global Food Animal Residue Avoidance Databank (gFARAD) program is a database intended to provide veterinarians with accurate guidance and information for food animal producers on the disposition of drugs or chemicals in animals prior to slaughter. In Canada, two regional centres participate in this program, based at the Western College of Veterinary Medicine in Saskatoon and the Faculté de médecine vétérinaire de l’Université de Montréal in St. Hyacinthe, Quebec. The services provided by gFARAD are intended to provide veterinarians with case-by-case information on withdrawal periods for veterinary drugs and on extra-label use of veterinary medications. This program also provides information regarding veterinary treatment for niche market food animals, exotic or minor use species in Canada for which no data exists regarding application of licensed drugs or appropriate withdrawal times.
INTERNATIONAL
Alliance for the Prudent Use of Antibiotics (APUA)
Hosted by Tufts University and guided by Dr. Stuart Levy, the Alliance for the Prudent Use of Antibiotics was created through the combined efforts of individuals, groups, institutions and countries with a shared concern for preserving the efficacy of antibiotic drugs and managing the health risks associated with the development of antimicrobial resistance. In existence for over 32 years, the mission of the Alliance remains unchanged: to improve and strengthen society’s defences against infectious diseases through improved antimicrobial availability and use.
Antimicrobial Resistance Management (ARM) Program
This on-going project based out of the University of Florida collects data from volunteer participating institutions to document trends in antimicrobial susceptibility in human patients and to determine associations between antibiotic use and rates of resistance in specific human pathogens. The ARM program gathers data from inpatient and outpatient isolates from six geographic regions across the United States. There are nearly four hundred participating institutions that provide information primarily related to Escherichia coli, Staphylococcus aureus and MRSA, Pseudomonas aeruginosa, Klebsiella pneumoniae and Streptococcus pneumoniae. The database enables comparisons in antimicrobial resistance trends over time, between hospitals, between geographic locations and between states and national data.
Global Antibiotic Resistance Partnership (GARP)
Funded by the Bill and Melinda Gates Foundation, the Global Antibiotic Resistance Partnership (GARP) is an initiative of the Center for Disease Dynamics, Economics & Policy at Resources for the Future in Washington, DC. The primary goal of this project is to develop strategies to delay the spread of antibiotic resistance—including strategies to reduce the need for antibiotics—in five low- and middle-income countries (India, Kenya, South Africa, Vietnam, and China).
3.11 Elements of an Optimal Surveillance Program for Monitoring Antibiotic Resistance
Examination of current surveillance programs for monitoring antibiotic utilization and antimicrobial resistance from various regions of the world reveals both strengths and shortcomings. There are, however, some key features that typify an optimal surveillance program. The Danish Integrated Antimicrobial Resistance Monitoring and Research Program (DANMAP) is often distinguished as the ‘gold standard’ for similar surveillance programs. This program represents a serious commitment on the part of the Danish government to support an ongoing longitudinal monitoring effort that collects data for human medical use and veterinary medical use of antibiotics and the incidence of antimicrobial resistance.
To optimize the value of surveillance efforts to monitor antibiotic use and antimicrobial resistance, programs need to be well-designed, adaptable, properly managed and able to detect significant shifts in microbial susceptibilities.123 Continuous surveillance of emergent pathogens and antimicrobial resistance is necessary in order to detect trends in real time and to characterize molecular mechanisms of resistance.124 For both national-level and international-level surveillance programs, there needs to be consistency in sampling and testing methods, pathogens, drugs, quality control measures and education, in order to compare trends in antimicrobial resistance and to assess effectiveness of prevention and control practices.125 Consideration of potential bias must be incorporated into the design and evaluation of antimicrobial resistant organism surveillance in terms of denominator data, case definitions, identification of all episodes that meet a case definition, sampling bias, reporting multiple occurrences of the same infectious case and bias related to laboratory analyses.126
For Canada the most important aspect of a surveillance program is that it be sustainable. While comprehensive programs such as DANMAP, NethMap and other Scandinavian programs are ideal, they are based on heavy commitment from a strong central government with almost total control over health care, prescriptions and agriculture. Canada’s power is more decentralized with much more provincial autonomy, especially concerning health and agriculture. Models such as EARS-Net and NARMS are much more realistic models for Canada, where a central organization sets case definitions and supports data collection and analysis, but regional authorities collect and control samples based on agreed-upon criteria. Use of pharmaceutical funds, while possibly functional in41 some systems (e.g. Britain), would not be acceptable for long-term funding of a surveillance system in Canada.
4. Program Evaluation, Comparison and Analysis
• An evaluative framework based on surveillance literature was applied to major Canadian AMR surveillance programs and compared with selected exemplar programs.
• Comparison of attributes and descriptions of pathogens of interest are presented in comparison tables for human AMR, human AMU, veterinary AMR and veterinary AMU.
• DanMAP, NethMap and the ECDC EARS-Net stand out as exemplar programs for AMR surveillance.
• All national Canadian programs are inherently limited in scope, and AMR surveillance data is overwhelmingly focused on hospitalized patients.
• The performance characteristic of “integration” was lacking for all Canadian programs.
• Utilization surveillance in Canada has only been reported for community AMU, derived from a proprietary data purchased from IMS Canada and reported nationally by CIPARS.
• Utilization surveillance of the exemplar national programs, DANMAP and NethMap, was rated as being quite comprehensive and complete.
• Animal AMR surveillance is most complete from Denmark and the European Union but the North American systems compare reasonably well. CIPARS and FoodNet Canada perform surveillance of enteric pathogen resistance in beef, pork and poultry production and retail meat. There are no data from veterinary microbiology for pathogens from diseased food animals or companion animals.
4.1 Program Ranking System
Selected attributes of major Canadian AMR surveillance programs and selected exemplar programs are displayed in Tables 4.1–4.4. The attribute structured evaluation tool (rubric) was developed based on core public health literature on the evaluation of surveillance,127 divided into system descriptors and performance characteristics. These attributes were compared with the elements of ideal surveillance identified in the Canadian expert interviews, confirming that all identified characteristics were encompassed. The program descriptions of pathogens of interest in surveillance were derived from review of the surveillance parameters of major programs and the pathogens of interest in AMR surveillance were identified by the experts interviewed. A colour based ranking system was applied.
• Red indicates that the surveillance system does not have the listed characteristic or does not perform the function.
• Yellow indicates partial presence or performance of the listed characteristic or function.
• Green indicates the system has or performs the characteristic or function within its known mandate and scope.
• Black indicates that the attribute or function lies outside the intended program.
The evaluation was applied by four team members (the principle investigators and project manager) with any discordant rankings resolved by iterative discussion and research on additional details on program attributes. The trans-disciplinary steering group also reviewed the final ranking tables, although there were some recent (April 2014) changes made to reflect intervening improvements in timeliness or new information. Descriptions of program characteristics for all programs on the tables are available in the systematic review discussion.
4.2 Results
(Click on images to enlarge.)
Table 4.1 : AMR – Human
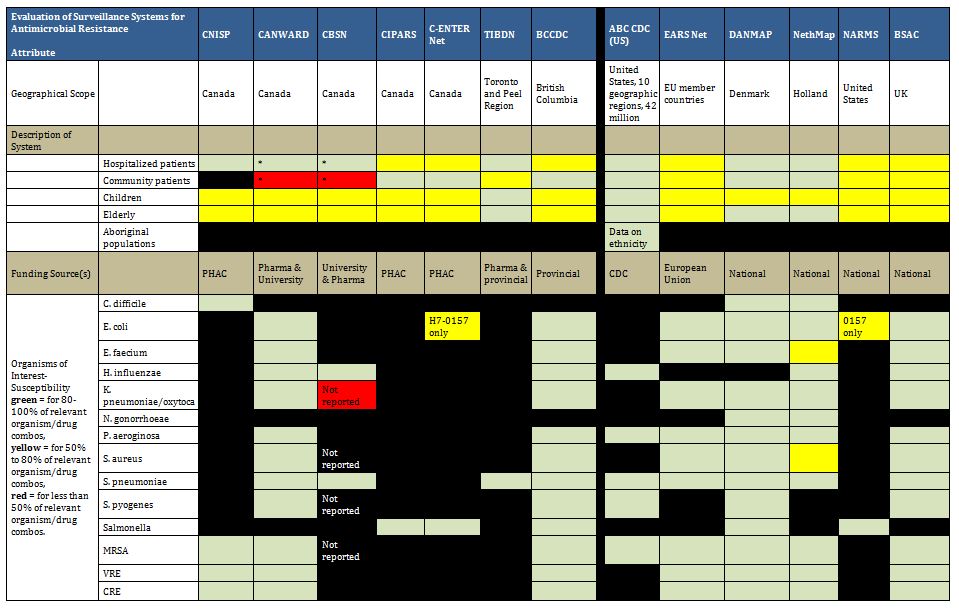
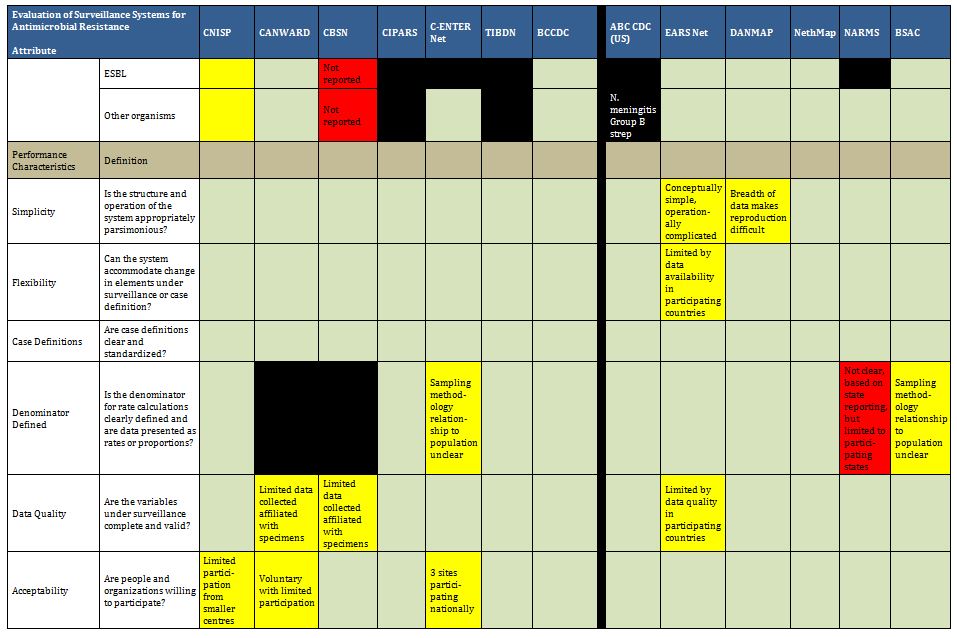
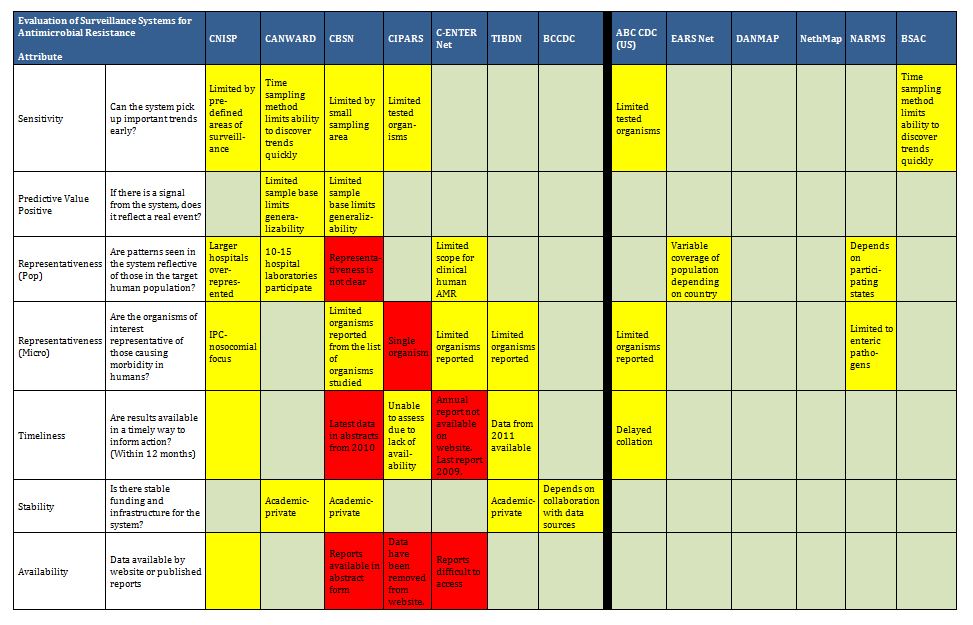
Table 4.2 : AMU – Human


Table 4.3: AMR – Veterinary

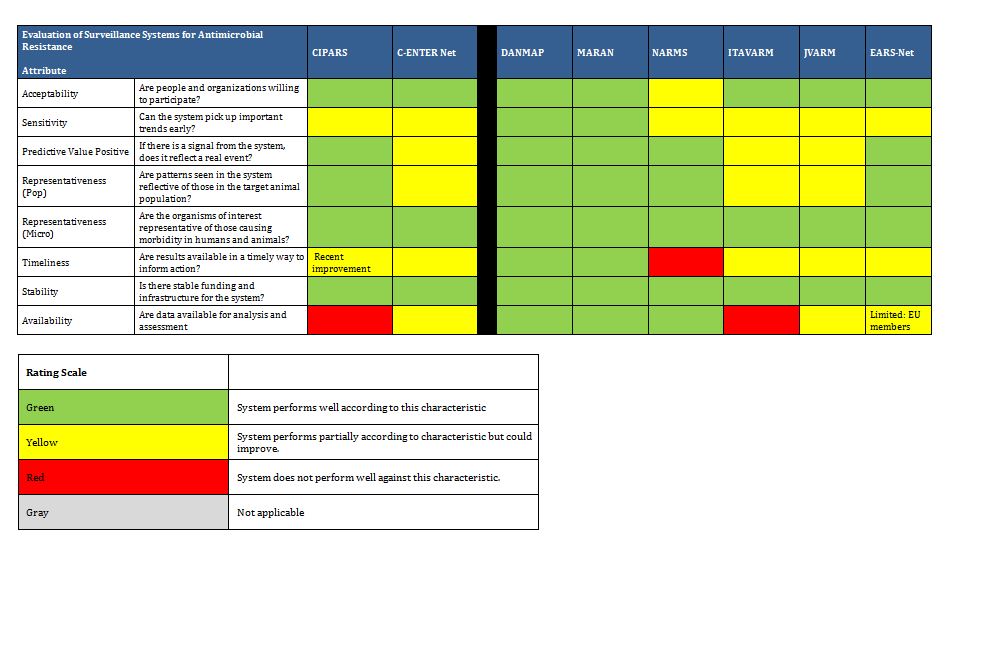
Table 4.4: AMU – Veterinary
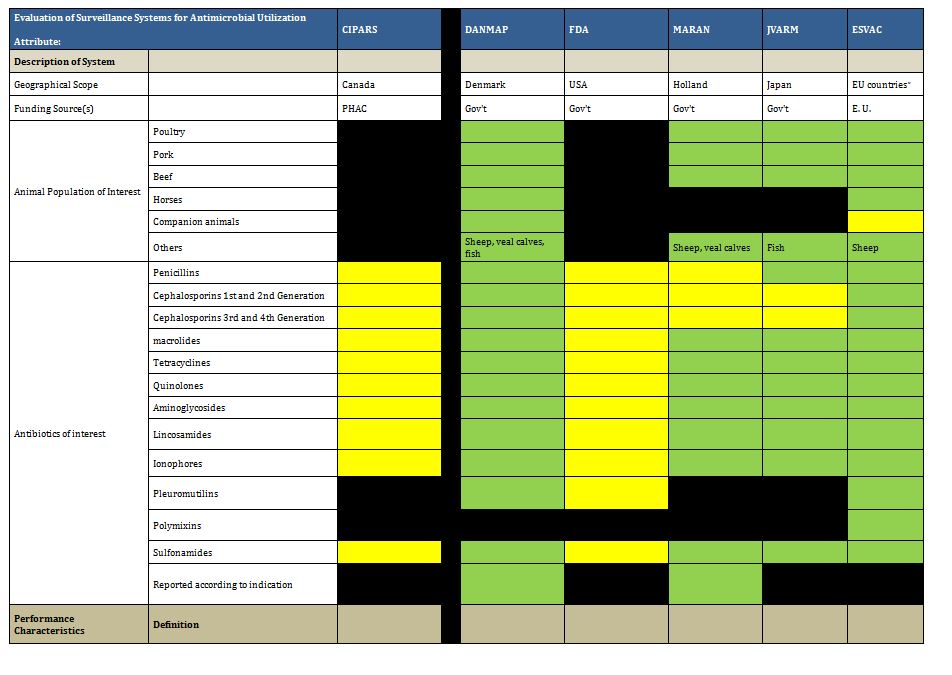
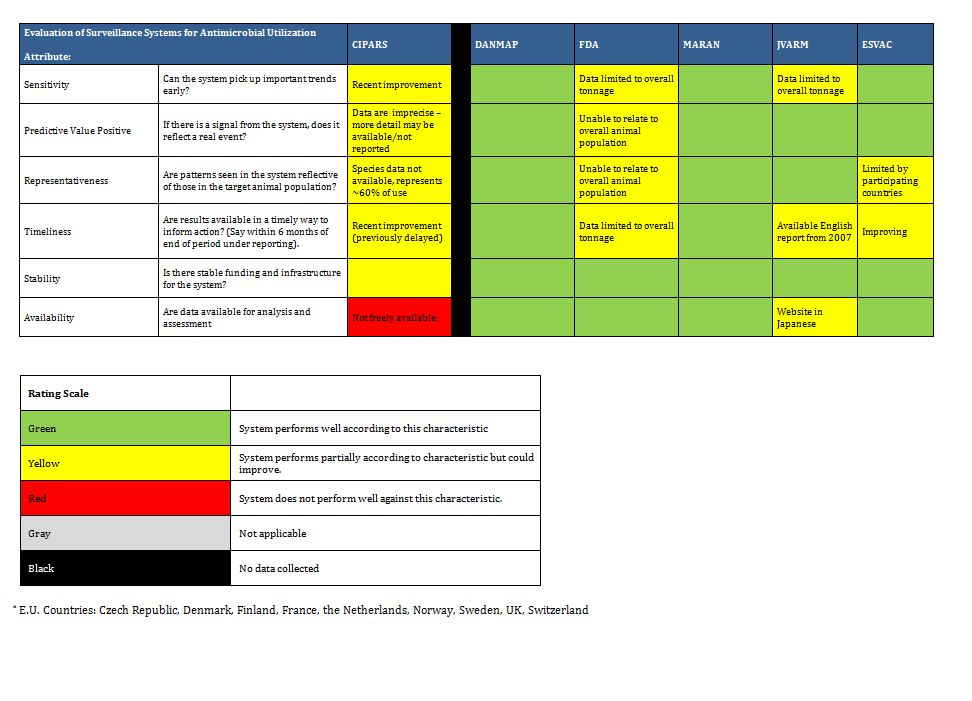
4.3 Discussion of Human AMR Surveillance Canadian Programs
Canadian Programs
The visualization of attributes in the preceding tables clearly demonstrates that key elements of AMR and AMU surveillance are not addressed on a national basis by current systems. All national Canadian programs are inherently limited in scope. This limitation is conferred in one of two ways:
1) By the patient population from which the resistance organisms under surveillance were collected.
In Canada, this is overwhelmingly focused on hospitalized patients. CNISP is dedicated to Infection Prevention and Control, and its surveillance is focused on nosocomially-acquired infections including some pathogens of AMR relevance. The longstanding, pharmaceutical company-supported CANWARD program performs prospective sampling of pathogens from clinical specimens submitted by network of tertiary hospital microbiology laboratories. Emergency room-based and clinic-based isolates would be considered community derived, but comprise a small subset that is not routinely analyzed and reported.
2) By the pathogens under surveillance, as a function of the program design and intent.
The CIPARS resistance surveillance is designed to evaluate the impact of antimicrobial use in food animal production, and thus is focused on Salmonella resistance surveillance in abattoirs, retail meat and eggs, and also surveillance of provincially submitted human salmonellosis isolates. This intersects somewhat with the FoodNet program, which performs enteric disease surveillance, including farm, water, and retail meat surveillance, as well as capturing enteric diseases that are notifiable to Public Health, at three sentinel communities across in Canada.
As evident from the tables, the organisms under national public health surveillance in Canada include AROs, which are followed as nosocomial pathogens by CNISP, and enteric pathogens, and are limited in number as well as scope.
The performance characteristic of timeliness has been a problem across public health and some academic surveillance programs, with challenges related to delays in the public availability of data posted on federal websites, and also related to public health and academic program reports being available only as presentations at scientific meetings and in scientific publications. Thus, they are subject to the timing of conferences and the processes of journal peer review and publication. There has been a notable increase in the volume of data and speed of posting available federally since late 2013, which resulted in changes to the original ranking in the rubric.
Finally, the performance characteristic of “integration” was lacking for all Canadian programs. In this rubric, “integration” was the term used to indicate that that the described system is a functioning part of a coordinated, public health based AMR surveillance system.
Our review of optimal systems resulted in the identification of the three following characteristics that were used to evaluate surveillance program integration: 1) explicit ties identifiable between surveillance system components; 2) a unified governance structure that is charged with coordination of surveillance programs as components of overall AMR surveillance for the jurisdiction, and determination of overall AMR surveillance needs and development; and 3) ties to AMR control programs, such as stewardship programs and policy development bodies that require AMR surveillance data to design interventions.
Strengths of Exemplar International Programs
The international exemplar surveillance programs identified by the literature review and by our expert interviews were all found to have stronger performance in the evaluation rubric. DANMAP and BSAC provided the most comprehensive (organism based) surveillance, and DANMAP and NethMap had the strongest community AMR surveillance. The EARS-Net program of the European Union had a shorter list of pathogens under surveillance but was notable for its effective coordination across multiple jurisdictions and provision of current, accessible, and useful (i.e. informs action and has impact) data.
Models of Development of AMR Surveillance
It is illustrative to expand upon the characteristics and evolution of the exemplar programs, focusing on DANMAP, NethMap and the ECDC EARS-Net, and compare them with the Canadian AMR surveillance systems.
Infrastructure
All have internal structures that acknowledge and explicitly tie together national jurisdictions in public health and agriculture-food safety. In smaller countries, surveillance programs were created by combining aspects of both of these as a surveillance program, under a new, crosscutting umbrella- e.g. DANMAP. Across the geographically broad and diverse European Union, where separate unit (country) level infrastructures had to be combined, participating units were given the tools required to submit data after appropriate agreements were developed, with ongoing reporting on data collection and results reinforcing compliance through generation of useful data reports and comparisons. In this case, the human AMR surveillance and the AMU surveillance, despite having analogous data collection tools, are handled separately. That is, we believe the agencies reporting different types of data do not necessarily directly connect within the country.
DATA SHARING
Exemplar programs also exhibit clarity of data availability and predictably timely reporting. This is accomplished through making data publicly available in regularly issued public reports, or in the case of the European Union, in published reports with interpretation and searchable databases – these are presented in a user-friendly fashion, with appropriate acknowledgement of the limitations of the data. Data sharing is meant to involve multiple stakeholders and the public, and is specifically not restricted to policymakers and only those involved in data collection and analysis. We note that the European Union members derive useful observations and impetus for positive change from seeing their own data contextualized and compared with other countries, and that this model may be appropriate across the provincial /territorial structures within Canada.
Program Creation and Evolution
The ECDC programs were essentially piloted in core countries then expanded with corresponding, simultaneous infrastructural development. An excerpt from the ECDC website describes it thus:
“The European Antimicrobial Resistance Surveillance System (EARSS), established in 1998, is the predecessor of the current EARS-Net.
“Following years of increasing concern for the occurrence and spread of antimicrobial resistance, the European Commission invited scientists, doctors and public health specialists to a ‘Microbial Threat Conference’ held in Denmark in September 1998. One conclusion of the conference was that a European surveillance system for antimicrobial resistance should be established.
“EARSS was initially funded by the European Commission’s Directorate General for Health and Consumer Affairs (DG SANCO) and the Dutch Ministry of Health, Welfare and Sports (RIVM). The network steadily grew and involved an increasing number of European countries. In 2001 a follow-up EU conference was held in Sweden and it was decided that all EU Member States should join the EARSS initiative.
“The EARSS network aimed to serve as a basis for an integrated public health strategy for containing antimicrobial resistance. In pursuing this, EARSS collaborated closely with other EU-funded projects e.g. the European Surveillance of Antimicrobial Consumption (ESAC) and Antibiotic Resistance Surveillance and Control in the Mediterranean Region (ARMed). EARSS also worked in partnership with the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and in particular with two of the society’s subcommittees: the European Committee on Antimicrobial Susceptibility testing (EUCAST) and the Study Group for Antimicrobial Resistance Surveillance (ESGARS).
“By January 1st, 2010, the administration and coordination of EARSS was transferred from RIVM to the European Centre for Disease Prevention and Control (ECDC). The network was renamed to ‘European Antimicrobial Resistance Surveillance Network (EARS-Net)’.”
Taken from: http://www.ecdc.europa.eu
In contrast, the exemplar nationally based programs –NethMap, and DANMAP – were created by the formation of national structures and the development of the scope and authority to carry out the surveillance, as appropriate for their respective countries. DANMAP was established by two Danish national ministries (see below), whereas NethMap was the result of a consortium of national professional societies (infectious diseases specialists, pharmacists and medical microbiologists) forming a nidus, which was then supported by the national health and wellness ministry, through the ECDC.
DANMAP
The Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP) was established by the Danish Ministry of Food, Agriculture and Fisheries and the Danish Ministry of Health in 1995. The objectives of the program are:
• to monitor the consumption of antimicrobial agents for food animals and humans;
• to monitor the occurrence of antimicrobial resistance in bacteria isolated from food animals, food of animal origin and humans;
• to study associations between antimicrobial consumption and antimicrobial resistance;
• to identify routes of transmission and areas for further research studies.
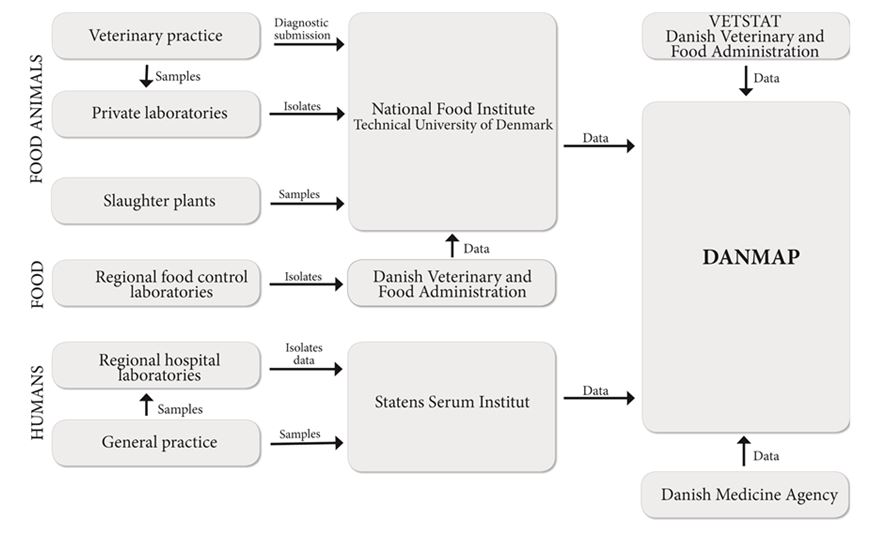
NethMap
NethMap was an initiative that started with professional societies in collaboration with and infrastructural support from federal public health.
“NethMap is a product of cooperative efforts of members of The Netherlands Society for Infectious Diseases, The Netherlands Society of Hospital Pharmacists and the Netherlands Society for Medical Microbiology. In 1996 the three societies created the Dutch Working Group on Antibiotic Policy, known as SWAB (Stichting Werkgroep Antibiotica Beleid).
SWAB, in collaboration with the RIVM, the National Institute for Public Health and the Environment of the Netherlands publishes the NethMap reports. SWAB is fully supported by a structural grant from the Ministry of Health, Welfare and Sports of the Netherlands. The information presented in NethMap is based on data from ongoing surveillance systems on the use of antimicrobial agents in human medicine and on the prevalence of resistance to relevant antimicrobial agents among medically important bacteria isolated from patients in the community and from patients admitted to hospitals.
Because of the multidisciplinary composition of SWAB, this foundation can be considered the Dutch equivalent of the Intersectoral Coordinating Mechanisms (ICMs), recommended by the European Union (2001), to control emerging antimicrobial resistance and promote rational antibiotic use. SWAB has started several major initiatives to achieve its goals. Among these are training programmes for the rational prescribing of antimicrobial drugs, development of evidence based prescription guidelines, the implementation of tailor made hospital guides for antibiotic prophylaxis and therapy and an integrated nationwide surveillance system for antibiotic use and antimicrobial resistance. These initiatives are corresponding well with the recommendations from the Dutch Council of Health Research (2001). Following these recommendations SWAB’s work was and still is made possible by structural funds provided by the Ministry of the Dutch Centre for Infectious Diseases Control (Centrum voor Infectieziektenbestrijding, CIb) in The National Institute of Public Health and the Environment (RIVM).
SWAB’s mission is to manage, limit and prevent the emergence of resistance to antimicrobial agents among medically important species of microorganisms in the Netherlands, thereby contributing to the quality of care in the Netherlands.”
[From: NethMap 2013 Report. Consumption of antimicrobial agents and antimicrobial resistance among medically important bacteria in the Netherlands. ]
North American (U.S. and Canada) Surveillance Programs – Room to Grow
American and Canadian surveillance programs identified by the literature review and by our expert interviews are observed to have some clear parallels.
1. The U.S. and Canada both have reasonable surveillance of enteric pathogen resistance in the context of agri-food (“from farm to fork”) through NARMS and CIPARS respectively.
2. The lack of Public Health surveillance of community level antimicrobial resistance in key pathogens is an easily identified weakness in both Canada and the U.S. Both countries have non-public health based community AMR surveillance via private company or pharma-academic partnerships.
a. In Canada, the CARA Alliance/CANWARD study has collected data prospectively from 10-15 academic hospital based laboratories, which send consecutive isolates for a set period of time yearly, based on an algorithm. While this generates useful data that attempts to address an important gap, this is a pharmaceutical-academic partnership, with no public health mandate or reporting. In addition, the proportion of truly community-based isolates is not clear in most reports from this data set. However, its wide geographic coverage and time trends make this program notable.
b. U.S. community based resistance indicators come from two sources:
The Surveillance Network (TSN), an “extensive collection of microbiological susceptibility test results used for surveillance of antimicrobial resistance. TSN is a private company that was established in 1994—its data set is quite extensive and to some extent mirrors the methodology of EARS-Net. In the absence of public health based information, researchers at the CDC, U.S. Food and Drug Administration (FDA), and Clinical and Laboratory, Standards Institute (CLSI) have drawn on its resources. Via direct technologic linkages, information is submitted on a daily basis from a selected network of some 300 clinical laboratories in nearly 200 zip codes. All laboratories adhere to CLSI standards. Participation is voluntary. Although certain states have no participating laboratories, TSN is considered representative at the national and census division levels.”
In addition, a population based public health data on a set of invasive pathogens is available through ABC Surveillance at the CDC, which engages all microbiology labs in select, large geographically defined areas. Although AMR evolution was not a primary driver in the creation of the system, resistance data are collected on the pathogens under surveillance.
3. The intersection of AMR and Infection Control Surveillance is observable in both countries.
a. In Canada, the CNISP Program collects valuable, longitudinal, population denominator-based surveillance of resistant pathogens of importance in nosocomially acquired infection.
b. Similarly, in the U.S., the National Healthcare Safety Network (NHSN) System of the CDC aggregates nosocomial infection data as a voluntary, hospital-based reporting system established to monitor hospital-acquired infections and guide the prevention efforts of infection control practitioners. The NHSN (formerly NNIS) system establishes a national risk-adjusted benchmark for nosocomial infection rates by using uniform case definitions and data-collection methods.
In both countries, the AMR data collected through infection prevention and control programs focus on nosocomial AROs, a group of pathogens that tend to be distinguished by 1) hospital acquisition of colonization or infection; and 2) distinct strains with distinct phenotypes, which are known to spread within hospitals and may be controllable through hygiene measures and isolation precautions. Thus, these organisms, when acquired by patients in health care environments, are already resistant to certain antibiotics, regardless of patients’ antibiotic exposures. This should be distinguished from the type of AMR that develops upon patient exposure to antimicrobials, either in the hospital or community. This latter type of resistance is a major data need, to guide appropriate antibiotic stewardship and guidelines development. (Another way of describing this is by differentiating nosocomial-resistant organisms which are “born” such as Staphylococcus aureus strains that are genetically methicillin resistant (regardless of antibiotic use in individual patients), and whose spread can be tracked through populations, versus resistance that is “made”, such as selection of resistant E. coli urinary tract infections in which antibiotic use in a patient increases the likelihood of their pathogen developing resistance. Overall antibiotic use in the community selects increasing community-wide resistance to that antibiotic.)
This latter kind of resistance tends to be complex and unpredictable, with the diverse inputs and selective pressures in the respective “microecosystems”. This type of AMR has not characteristically been tracked by IPC programs, whose mandate is focused on hospital-based spread of defined resistant bacterial strains in order to develop effective control programs.
In the Canadian context, it is important to acknowledge the need for nosocomial ARO tracking, and define how this can differ from community and hospital based AMR surveillance as tied more closely to antibiotic utilization pressures. An optimal system of surveillance will benefit from both data sources, and areas of overlap may be exploited to allow fruitful collaboration and best use of available infrastructure and federal expertise.
4.4 Human AMU Surveillance
Canadian Programs
Utilization surveillance in Canada has only been performed and reported for community AMU, derived from a proprietary data purchased from IMS Canada and reported nationally by CIPARS. These reports provide useful and important data but until very recently, timeliness of reports and thus ability to address emerging problems was limited. Further use of these data with more extensive sub-analysis and integration with provincial public health and antimicrobial stewardship initiatives, would improve the utility. Although it was understood that hospital purchasing data are under analysis by CIPARS and that a subset of the CNISP hospitals are engaged in hospital utilization data collection, no reports were available at the time of writing.
On a provincial level, the BCCDC deserves mention as a potential Canadian model system. The BCCDC reports demonstrate the utility of non-proprietary data and linked data systems. There is a capability for integrating utilization data from PharmaNet, a provincial system which tracks all dispensed prescriptions and resistance data from sources previous described (predominantly community data). This allows development of responsive provincial guidelines, as well as the possibility of prescriber based audit and feedback, which has potential to be a powerful tool in improving utilization.
International Programs
Utilization surveillance of the exemplar national programs DANMAP and NethMap was rated as being quite comprehensive and complete, and similar to the AMR surveillance. The utilization data collected by the ECDC for the European Union was seen as useful and timely but had a relatively restricted list of antibiotics under surveillance. There were no U.S. antibiotic utilization surveillance programs for direct comparison.
4.5 Animal AMR-AMU Surveillance
Canadian Programs
CIPARS and FoodNet Canada perform surveillance of enteric pathogen resistance in beef, pork and poultry production and retail meat. There are no data from veterinary microbiology (pathogens from diseased food or companion animals). CIPARS reports some utilization data based on purchasing, which is not available in all reporting years. The current oversight and control of animal antibiotics in Canada imposes significant limitations in being able to track and assess animal antibiotic utilization, with importation loopholes that are difficult to track without regulatory authority (the Own Use provision, and importation of API (active pharmaceutical ingredients).
International Programs
Animal AMR surveillance, as seen in table 4.3, is most complete from Denmark and the European Union, but the North American systems still compare reasonably well. The surveillance of antibiotic use in animals is much more complex given the multiple classifications of use, variety of users and prescribers, and various purchasing/regulatory systems that can exist.
DANMAP and ESVAC have the most comprehensive utilization data across animal populations and across antimicrobials tracked. It is noteworthy that the Danish approach to tracking and regulation of animal antibiotic use has resulted in 21% reduction of antibiotic use in pigs and is held as a standard.128 Thus, a more detailed description is provided. The Danish VetStat program collected data from veterinarians, pharmacies, and feed mills since 2000, monitoring prescription medicine in production animals as the use of coccidiostatics, with data collected at the farm level including animal species, age of animal, disease, farm identification number, veterinarians’ number, drug identification number, amount of medicine, and date for use of medicine. Today VetStat enables authorities to assess usage patterns at the level of the individual herd and individual prescriber. Furthermore, many veterinarians use VetStat daily as a tool in relation to their service for their clients (farmers). Because all data are converted to defined animal daily doses (ADDs) it is possible to compare the use of antibiotics on one farm with a similar average for the whole country. In 2010 the Danish Veterinary and Food Authority (DVFA) introduced the “Yellow Card Initiative” based on VetStat, in which threshold limits for antimicrobial consumption in pigs were established; if thresholds are exceeded, the DVFA may issue an order or injunction (the yellow card) compelling the owner of the holding, in collaboration with the veterinary practitioner, to reduce the antimicrobial consumption in the holding below the threshold limits within nine months.
The total use of antimicrobials in swine has been reduced by 21 per cent in Denmark, following the introduction of the Yellow Card Initiative, when comparing national data on usage for the years 2009 and 2011, respectively.
4.6 Summary: Outstanding Issues and Next Steps
This project has described Canadian and international programs of surveillance for AMR and AMU. In Chapter 3, eleven programs in Canada were described in terms of their coverage, responsibilities and reporting. Chapter 4 summarized how well select programs meet the criteria for process and content considered “best” by expert reviewers. Where possible, Chapter 4 also provided information about the infrastructure and the funding models for those programs. Although all programs could be strengthened in some ways, there are good examples which Canada, with its federated systems, can adapt for surveillance of AMR and AMU in this country.
According to our criteria, DANMAP and BSAC provide the most comprehensive surveillance in terms of selected organisms, and DANMAP and NethMap are the strongest programs found in terms of community-level surveillance of AMR. Although covering a short list of pathogens, the European Union EARS-Net program effectively coordinates surveillance across many jurisdictions, collects data from both human- and animal-related systems, and makes its current data available As noted, these three programs have sustained infrastructure and also have mechanisms and processes to share data publicly through timely and regular reporting.
In comparison, Canada’s surveillance systems have not yet achieved the comprehensiveness, integration and coordination of the four strongest European surveillance programs. As noted, surveillance of enteric pathogen resistance in agri-food is good, as is surveillance of nosocomial AROs in hospitals, but there is still a gap in comprehensive surveillance for human specimens in the community. Surveillance of human AMU in the community is hampered by delays in the release of data and reports and the current inability to make comparisons among the provinces. The development of national data-sharing agreements, collaborative development of agreed-upon antibiogram processes, definitions, and technical infrastructure would enable better data collation and comparison. Support for distinguishing in-hospital from community isolates would complement that data available on hospital-driven AMR, and provide for anonymized line listed data by isolate.
Reviewing the recommendations that emerged in previous reports (Appendix F), it is evident that these issues have been identified before and require thoughtful action to address them as soon as feasible. These previous reports reflected on the need for dedicated federal leadership infrastructure, full engagement of the provinces and territories, and shared responsibility to create or enhance the infrastructure needed in all the jurisdictions. Meeting these three criteria will enable national collaboration and coordination that includes the provincial and territorial ministries for health and agriculture (including veterinary services). Additionally, the earlier recommendations have noted the need for benchmarks which can be monitored with regular and timely release of data. Our analysis of programs in this report supports those recommendations.
The solutions to these problems build upon the existing programs and surveillance systems. Canada can look to the European models described above to develop a coordinated effort among federal, provincial and territorial governments that can enhance the mechanisms and processes in place at sentinel sites and in hospitals, expanding them to include AMU and AMR surveillance in the community including long term care facilities, laboratories and in agriculture.
5. Conclusion
We suggest that the status of Canadian surveillance of Antimicrobial Resistance and Antimicrobial Utilization is at a critical point, as AMR is becoming a preeminent worldwide threat to the public health. Current systems do not track evolving AMR in the community or hospital and thus cannot support development of meaningful responses through better stewardship of antimicrobial use in community or hospital settings, or in veterinary and in food animal production. We have limited information on antibiotic use in many settings, and thus establishing controls and evaluating the impact of intervention becomes impossible. Simply put, we need more comprehensive data to be able to analyze the problem, develop a response, and then look for effectiveness of that response.
We must acknowledge that the weaknesses and gaps identified in this report exist for a reason: the challenge of national surveillance is a truly difficult problem in our health care system, with even the “ownership” of the problem, and the response, seeming unclear. The delivery of health care is primarily an area of provincial jurisdiction, but clearly public health surveillance requires national coordination, to allow integration of the most appropriate data into a national AMR-AMU picture.
The process of review, analysis, comparison, and discussions during our companion trans-Canadian, transdisciplinary expert survey helped focus the recommendations presented on two key issues: who is accountable, and how to model the evolution to comprehensive surveillance.
By building up from the strongest surveillance components we have at a national level, CIPARS and CNISP, we would hope use the successes of these programs in establishing national frameworks and collaborations. Bringing these two programs, which represent very different areas of AMR-AMU surveillance together could centralize existing expertise, and start to integrate government AMR portfolios in a direction towards “One Health.”
The area of accountability which has been brought forth prominently here is the importance of provincial and territorial governments and public health agencies in supporting and creating a framework that can benefit all, under a public health mandate. As the need for useful data has become more apparent, we have seen the creation of organism-based, geography-based, and short term surveillance projects to fill the gaps. But what is required is a strong collaborative body to bring the groups together to refine exactly what we need to collect and then share to inform Canadian public health. Potentially the most important task ahead seems the most prosaic – confirming Provincial and Territorial support for AMR–AMU surveillance coordination, and forging agreements on areas such as: which AMR and AMU data are to be prioritized for collection, the development of collaborations to further create and then share needed processes, with division of tasks based existing programs and expertise and parameters for data reporting.
For the potential benefits, the monetary investments needed are relatively small, but the goodwill and collaboration needed are large. However, history may judge us harshly should we fail to protect the effectiveness of antimicrobials in animal and human health.
References
1. Fleming A. Penicillin. Nobel Lectures Amsterdam: Elsevier Publishing Company; 1945.
2. Spellberg B, Bartlett JG, Gilbert DN. The Future of Antibiotics and Resistance. N Engl J Med 2013 01/24; 2014/05;368(4):299-302. 3. Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013. U S Department of Health and Human Services 2013 April 23.
4. World Health Organization. Antibiotic resistance – a threat to global health security. 2014; Available at: http://www.who.int/drugresistance/activities/wha66_side_event/en/. Accessed 05/25, 2014.
5. European Centre for Disease Prevention and Control. Summary of the latest data on antibiotic resistance in the European Union. 2013.
6. World Economic Forum. Global Risks 2013, Eighth Edition: An initiative of the Risk Response Network. 2013.
7. Mindlin SZ, Soina VS, Ptrova MA, Gorlenko Z. Isolation of antibiotic resistance bacterial strains from East Siberia permafrost sediments. Genetika 2008 Jan;44(1):36-44.
8. Baquero F, Alvarez-Ortega C, Martinez JL. Ecology and evolution of antibiotic resistance. Environmental Microbiology Reports 2009 12/01;1(6):469-476.
9. Wright GD. Antibiotic resistance in the environment: a link to the clinic? Curr Opin Microbiol 2010 Oct;13(5):589-594.
10. Martin I, Jayaraman G, Wong T, Liu G, Gilmour M, Canadian Public Health Laboratory Network. Trends in antimicrobial resistance in Neisseria gonorrhoeae isolated in Canada: 2000-2009. Sex Transm Dis 2011 Oct;38(10):892-898.
11. Filice GA, Nyman JA, Lexau C, Lees CH, Bockstedt LA, Como-Sabetti K, et al. Excess costs and utilization associated with methicillin resistance for patients with Staphylococcus aureus infection. Infect Control Hosp Epidemiol 2010 Apr;31(4):365- 373.
12. Cosgrove SE. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis 2006 Jan 15;42 Suppl 2:S82-9.
13. Pagel SW, Gautier P. Use of antimicrobial agents in livestock. Rev Sci Tech 2012 Apr;31(1):145-188.
14. Shea KM. Antibiotic Resistance: What is the impact of agricultural uses of antibiotics on children’s health? Pediatrics 2003 07/01;112:253-258.
15. Witte W. Ecological impact of antibiotic use in animals on different complex microflora: environment. Int J Antimicrob Agents 2000 May;14(4):321-325.
16. Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob Agents Chemother 2011 Mar;55(3):1274-1278.
17. One Health Initiative. About the One Health Initiative. Available at: http://www.onehealthinitiative.com/about.php. Accessed 05/06, 2014.
18. Palmer G, Call D. Antimicrobial resistance: a global public health challenge requiring a global one health strategy. Commentary, Institute of Medicine 2013 February 7.
19. World Health Organization. WHO Global Strategy for Containment of Antimicrobial Resistance. World Health Organization 2001.
20. Austin DJ, Kristinsson KG, Anderson RM. The relationship between the volume of antimicrobial consumption in human communities and the frequency of resistance. Proc Natl Acad Sci U S A 1999 Feb 2;96(3):1152-1156.
21. National Center for Emerging and Zoonotic Infectious Diseases. Strategic Plan 2012- 2017. Centers for Disease Control, CDC 2012;CS234429-A.
22. Bager F. DANMAP: monitoring antimicrobial resistance in Denmark. Int J Antimicrob Agents 2000 May;14(4):271-274.
23. Johnson AP, Davies J, Guy R, Abernethy J, Sheridan E, Pearson A, et al. Mandatory surveillance of methicillin-resistant Staphylococcus aureus (MRSA) bacteraemia in England: the first 10 years. J Antimicrob Chemother 2012 Apr;67(4):802-809.
24. Lafaurie M, Porcher R, Donay JL, Touratier S, Molina JM. Reduction of fluoroquinolone use is associated with a decrease in methicillin-resistant Staphylococcus aureus and fluoroquinolone-resistant Pseudomonas aeruginosa isolation rates: a 10 year study. J Antimicrob Chemother 2012 Apr;67(4):1010-1015.
25. Soeters HM, von Gottberg A, Cohen C, Quan V, Klugman KP. Trimethoprimsulfamethoxazole prophylaxis and antibiotic nonsusceptibility in invasive pneumococcal disease. Antimicrob Agents Chemother 2012 Mar;56(3):1602-1605.
26. Kaplan SL, Mason EO, Jr. Management of infections due to antibiotic-resistant Streptococcus pneumoniae. Clin Microbiol Rev 1998 Oct;11(4):628-644.
27. Nuermberger EL, Bishai WR. Antibiotic resistance in Streptococcus pneumoniae: what does the future hold? Clin Infect Dis 2004 May 15;38 Suppl 4:S363-71.
28. Starr JA, Fox GW, Clayton JK. Streptococcus pneumoniae: An Update on Resistance Patterns in the United States. Journal of Pharmacy Practice 2008 10/01;21(5):363-370.
29. Linares J, Ardanuy C, Pallares R, Fenoll A. Changes in antimicrobial resistance, serotypes and genotypes in Streptococcus pneumoniae over a 30-year period. Clin Microbiol Infect 2010 May;16(5):402-410.
30. Vasoo S, Singh K, Hsu LY, Chiew YF, Chow C, Lin RT, et al. Increasing antibiotic resistance in Streptococcus pneumoniae colonizing children attending day-care centres in Singapore. Respirology 2011 Nov;16(8):1241-1248.
31. Goossens H, Ferech M, Coenen S, Stephens P, European Surveillance of Antimicrobial Consumption Project Group. Comparison of outpatient systemic antibacterial use in 2004 in the United States and 27 European countries. Clin Infect Dis 2007 Apr 15;44(8):1091- 1095.
32. Goossens H. Antibiotic consumption and link to resistance. Clin Microbiol Infect 2009 Apr;15 Suppl 3:12-15.
33. Malhotra-Kumar S, Lammens C, Coenen S, Van Herck K, Goossens H. Effect of azithromycin and clarithromycin therapy on pharyngeal carriageof macrolide-resistant streptococci in healthy volunteers: a randomised, double-blind, placebo-controlled study. Lancet 2007 Feb 10;369(9560):482-490.
34. Cornaglia G, Hryniewicz W, Jarlier V, Kahlmeter G, Mittermayer H, Stratchounski L, et al. European recommendations for antimicrobial resistance surveillance. Clin Microbiol Infect 2004 Apr;10(4):349-383.
35. Finch R. Antibiotic resistance–from pathogen to disease surveillance. Clin Microbiol Infect 2002 Jun;8(6):317-320.
36. Health Canada and The Canadian Infectious Disease Society. Controlling antimicrobial resistance. An integrated action plan for Canadians. Can Commun Dis Rep 1997 Nov;23 Suppl 7:i-iv, 1-32, i-iv, 1-32.
37. Canadian Committee on Antibiotic Resistance. Report on the 2002 National Policy Conference on Antimicrobial Stewardship. 2004.
38. Prescott JF, Dowling PM. Agriculture’s role in managing antimicrobial resistance: conference report. Can Vet J 2000 Mar;41(3):191-197.
39. Prescott JF, Szkotnicki J, McClure JT, Reid-Smith RJ, Leger DF. Conference report: antimicrobial stewardship in Canadian agriculture and veterinary medicine. How is Canada doing and what still needs to be done? Can Vet J 2012 Apr;53(4):402-407.
40. Health Canada and The Canadian Infectious Disease Society. Controlling antimicrobial resistance. An integrated action plan for Canadians. Can Commun Dis Rep 1997 Nov;23 Suppl 7:i-iv, 1-32, i-iv, 1-32.
41. Canadian Committee on Antibiotic Resistance. The Pan-Canadian Stakeholder Consultations on Antimicrobial Resistance. Public Health Agency of Canada 2009 September:3-62.
42. Wilson J, Conly J, Wong T, Jayaraman G, Sargeant J, Papadopoulos A, et al. Strategies to control community-associated antimicrobial resistance among enteric bacteria and methicillin-resistant Staphylococcus aureus in Canada – executive summary. Can J Infect Dis Med Microbiol 2010 Fall;21(3):133-137.
43. Atkins D, Eccles M, Flottorp S, Guyatt GH, Henry D, Hill S, et al. Systems for grading the quality of evidence and the strength of recommendations I: critical appraisal of existing approaches The GRADE Working Group. BMC Health Serv Res 2004 Dec 22;4(1):38.
44. Balshem H, Helfand M, Schunemann HJ, Oxman AD, Kunz R, Brozek J, et al. GRADE guidelines: 3. Rating the quality of evidence. J Clin Epidemiol 2011 Apr;64(4):401-406.
45. Canfield SE, Dahm P. Rating the quality of evidence and the strength of recommendations using GRADE. World J Urol 2011 Jun;29(3):311-317.
46. Neu HC, Duma RJ, Jones RN, McGowan JE, Jr, O’Brien TF, Sabath LD, et al. Antibiotic resistance. Epidemiology and therapeutics. Diagn Microbiol Infect Dis 1992 Feb;15(2 Suppl):53S-60S.
47. Morris AK, Masterton RG. Antibiotic resistance surveillance: action for international studies. J Antimicrob Chemother 2002 Jan;49(1):7-10.
48. O’Brien TF, Stelling J. Integrated Multilevel Surveillance of the World’s Infecting Microbes and Their Resistance to Antimicrobial Agents. Clin Microbiol Rev 2011 Apr;24(2):281-295.
49. Silley P, Simjee S, Schwarz S. Surveillance and monitoring of antimicrobial resistance and antibiotic consumption in humans and animals. Rev Sci Tech 2012 Apr;31(1):105- 120.
50. Zoutman DE, Ford BD. The relationship between hospital infection surveillance and control activities and antibiotic-resistant pathogen rates. Am J Infect Control 2005 Feb;33(1):1-5.
51. Ofner-Agostini M, Varia M, Johnston L, Green K, Simor A, Amihod B, et al. Infection control and antimicrobial restriction practices for antimicrobial-resistant organisms in Canadian tertiary care hospitals. Am J Infect Control 2007 Nov;35(9):563-568.
52. Adam HJ, Louie L, Watt C, Gravel D, Bryce E, Loeb M, et al. Detection and characterization of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates in Canada: results from the Canadian Nosocomial Infection Surveillance Program, 1995-2006. Antimicrob Agents Chemother 2010 Feb;54(2):945-949.
53. Hoban DJ, Zhanel GG. Introduction to the CANWARD study (2007-11). J Antimicrob Chemother 2013 May;68 Suppl 1:i3-5.
54. Hoban DJ, Zhanel GG. Introduction to the CANWARD Study (2007-2009). Diagn Microbiol Infect Dis 2011 Mar;69(3):289-290.
55. McCracken M, Mataseje LF, Loo V, Walkty A, Adam HJ, Hoban DJ, et al. Characterization of Acinetobacter baumannii and meropenem-resistant Pseudomonas aeruginosa in Canada: results of the CANWARD 2007-2009 study. Diagn Microbiol Infect Dis 2011 Mar;69(3):335-341.
56. Simner PJ, Zhanel GG, Pitout J, Tailor F, McCracken M, Mulvey MR, et al. Prevalence and characterization of extended-spectrum beta-lactamase- and AmpC beta-lactamaseproducing Escherichia coli: results of the CANWARD 2007-2009 study. Diagn Microbiol Infect Dis 2011 Mar;69(3):326-334.
57. Zhanel GG, Adam HJ, Low DE, Blondeau J, Decorby M, Karlowsky JA, et al. Antimicrobial susceptibility of 15,644 pathogens from Canadian hospitals: results of the CANWARD 2007-2009 study. Diagn Microbiol Infect Dis 2011 Mar;69(3):291-306.
58. Adam HJ, Karlowsky JA, Nichol KA, Gilmour MW, Hoban DJ, Embree J, et al. Baseline epidemiology of Streptococcus pneumoniae serotypes in Canada prior to the introduction of the 13-valent pneumococcal vaccine. Microb Drug Resist 2012 Apr;18(2):176-182.
59. Karlowsky JA, Walkty AJ, Adam HJ, Baxter MR, Hoban DJ, Zhanel GG. Prevalence of antimicrobial resistance among clinical isolates of Bacteroides fragilis group in Canada in 2010-2011: CANWARD surveillance study. Antimicrob Agents Chemother 2012 Mar;56(3):1247-1252.
60. Walkty A, Baxter M, Adam H, Karlowsky JA, Lagace-Wiens P, Hoban DJ, et al. Antimicrobial susceptibility of Pseudomonas aeruginosa isolates obtained from patients in Canadian hospitals: CANWARD 2008-2011. Diagn Microbiol Infect Dis 2012 Aug;73(4):361-364.
61. Denisuik AJ, Lagacé-Wiens PR, Pitout JD, Mulvey MR, Simner PJ, Tailor F, et al. Molecular epidemiology of extended-spectrum beta-lactamase-, AmpC beta-lactamaseand carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolated from Canadian hospitals over a 5 year period: CANWARD 2007-11. J Antimicrob Chemother 2013 May;68 Suppl 1:i57-65.
62. Karlowsky JA, Adam HJ, Desjardins M, Lagace-Wiens PR, Hoban DJ, Zhanel GG, et al. Changes in fluoroquinolone resistance over 5 years (CANWARD 2007-11) in bacterial pathogens isolated in Canadian hospitals. J Antimicrob Chemother 2013 May;68 Suppl 1:i39-46.
63. Zelenitsky SA, Rubinstein E, Ariano RE, Zhanel GG, Canadian Antimicrobial Resistance Alliance. Integrating pharmacokinetics, pharmacodynamics and MIC distributions to assess changing antimicrobial activity against clinical isolates of Pseudomonas aeruginosa causing infections in Canadian hospitals (CANWARD). J Antimicrob Chemother 2013 May;68 Suppl 1:i67-72.
64. Karlowsky JA, Adam HJ, Decorby MR, Lagace-Wiens PR, Hoban DJ, Zhanel GG. In vitro activity of ceftaroline against gram-positive and gram-negative pathogens isolated from patients in Canadian hospitals in 2009. Antimicrob Agents Chemother 2011 Jun;55(6):2837-2846.
65. Walkty A, DeCorby M, Nichol K, Karlowsky JA, Hoban DJ, Zhanel GG. In vitro activity of colistin (polymyxin E) against 3,480 isolates of gram-negative bacilli obtained from patients in Canadian hospitals in the CANWARD study, 2007-2008. Antimicrob Agents Chemother 2009 Nov;53(11):4924-4926.
66. Karlowsky JA, Adam HJ, Poutanen SM, Hoban DJ, Zhanel GG, Canadian Antimicrobial Resistance Alliance (CARA). In vitro activity of dalbavancin and telavancin against staphylococci and streptococci isolated from patients in Canadian hospitals: results of the CANWARD 2007-2009 study. Diagn Microbiol Infect Dis 2011 Mar;69(3):342-347.
67. Walkty A, DeCorby M, Lagace-Wiens PR, Karlowsky JA, Hoban DJ, Zhanel GG. In vitro activity of ceftazidime combined with NXL104 versus Pseudomonas aeruginosa isolates obtained from patients in Canadian hospitals (CANWARD 2009 study). Antimicrob Agents Chemother 2011 Jun;55(6):2992-2994.
68. Walkty A, Adam HJ, Laverdière M, Karlowsky JA, Hoban DJ, Zhanel GG, et al. In vitro activity of ceftobiprole against frequently encountered aerobic and facultative Grampositive and Gram-negative bacterial pathogens: results of the CANWARD 2007-2009 study. Diagn Microbiol Infect Dis 2011 Mar;69(3):348-355.
69. Hammerum AM, Heuer OE, Emborg HD, Bagger-Skjot L, Jensen VF, Rogues AM, et al. Danish integrated antimicrobial resistance monitoring and research program. Emerg Infect Dis 2007 Nov;13(11):1632-1639.
70. DANMAP. About DANMAP -. 2013; Available at: http://www.danmap.org/About%20Danmap.aspx. Accessed 05/07, 2014.
71. DANMAP. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. 2009 Report. 2009.
72. DANMAP. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. 2011 Report. 2011.
73. DANMAP. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. 2010 Report. 2010 September.
74. Aarestrup FM, Bager F, Jensen NE, Madsen M, Meyling A, Wegener HC. Resistance to antimicrobial agents used for animal therapy in pathogenic-, zoonotic- and indicator bacteria isolated from different food animals in Denmark: a baseline study for the Danish Integrated Antimicrobial Resistance Monitoring Programme (DANMAP). APMIS 1998 Aug;106(8):745-770.
75. Bager F, Aarestrup FM, Jensen NE, Madsen M, Meyling A, Wegener HC. Design of a system for monitoring antimicrobial resistance in pathogenic, zoonotic and indicator bacteria from food animals. Acta Vet Scand Suppl 1999;92:77-86.
76. Monnet D, Hemborg H, Andersen S, Scholler C, Sorensen T, Bager F. Surveillance of antimicrobial resistance in Denmark. Euro Surveill 2000 12;5(12):129-132.
77. Abatih EN, Emborg HD, Jensen VF, Lo Fo Wong DM, Ersboll AK. Regional, seasonal, and temporal variations in the prevalence of antimicrobial-resistant Escherichia coli isolated from pigs at slaughter in Denmark (1997-2005). Foodborne Pathog Dis 2009 Apr;6(3):305-319.
78. Skjot-Rasmussen L, Ethelberg S, Emborg HD, Agerso Y, Larsen LS, Nordentoft S, et al. Trends in occurrence of antimicrobial resistance in Campylobacter jejuni isolates from broiler chickens, broiler chicken meat, and human domestically acquired cases and travel associated cases in Denmark. Int J Food Microbiol 2009 May 31;131(2-3):277-279.
79. Vieira AR, Houe H, Wegener HC, Lo Fo Wong DM, Bodker R, Emborg HD. Spatial scan statistics to assess sampling strategy of antimicrobial resistance monitoring program. Foodborne Pathog Dis 2009 Jan-Feb;6(1):15-21.
80. Adriaenssens N, Coenen S, Versporten A, Muller A, Minalu G, Faes C, et al. European Surveillance of Antimicrobial Consumption (ESAC): outpatient antibiotic use in Europe (1997-2009). J Antimicrob Chemother 2011 12/01;66:vi3-vi12.
81. European Centre for Disease Prevention and Control. The European Antibiotic Resistance Surveillance Network (EARS-Net) Reporting Protocol, Version 3. 2013.
82. MARAN. Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in The Netherlands In 2004. 2005. 83. MARAN. Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands in 2010/2011. 2012.
84. Molstad S, Cars O, Struwe J. Strama. A Swedish working model for containment of antibiotic resistance. Euro Surveill 2008 Nov 13;13(46):19041.
85. Struwe J. Fighting antibiotic resistance in Sweden—past, present and future. Wien Klin Wochenschr 2008;120(9-10):268-279.
86. Nissinen A, Jarvinen H, Liimatainen O, Jahkola M, Huovinen P. Antimicrobial resistance in Neisseria gonorrhoeae in Finland, 1976 to 1995. The Finnish Study Group For Antimicrobial Resistance. Sex Transm Dis 1997 Nov;24(10):576-581.
87. Manninen R, Auvinen H, Huovinen P, The Finnish Study Group for Antimicrobial Resistance (FiRe). Resistance to second- and third-generation cephalosporins among Escherichia coli and Klebsiella species is rare in Finland. Clin Microbiol Infect 1997 Aug;3(4):408-413.
88. Pihlajamaki M, Kotilainen P, Kaurila T, Klaukka T, Palva E, Huovinen P, et al. Macrolide-resistant Streptococcus pneumoniae and use of antimicrobial agents. Clin Infect Dis 2001 Aug 15;33(4):483-488.
89. Karpanoja P, Nyberg ST, Bergman M, Voipio T, Paakkari P, Huovinen P, et al. Connection between trimethoprim-sulfamethoxazole use and resistance in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis. Antimicrob Agents Chemother 2008 Jul;52(7):2480-2485.
90. Kanerva M, Ollgren J, Hakanen AJ, Lyytikainen O. Estimating the burden of healthcareassociated infections caused by selected multidrug-resistant bacteria Finland, 2010. Antimicrob Resist Infect Control 2012 Oct 19;1(1):33
91. Morgan DJ, Okeke IN, Laxminarayan R, Perencevich EN, Weisenberg S. Nonprescription antimicrobial use worldwide: a systematic review. Lancet Infect Dis 2011 Sep;11(9):692-701.
92. White AR. The British Society for Antimicrobial Chemotherapy Resistance Surveillance Project: a successful collaborative model. J Antimicrob Chemother 2008 11/01;62:ii3- ii14.
93. Miyakis S, Pefanis A, Tsakris A. The challenges of antimicrobial drug resistance in Greece. Clin Infect Dis 2011 Jul 15;53(2):177-184.
94. Vatopoulos AC, Kalapothaki V, Legakis NJ. Bacterial resistance to ciprofloxacin in Greece: results from the National Electronic Surveillance System. Greek Network for the Surveillance of Antimicrobial Resistance. Emerg Infect Dis 1999 May-Jun;5(3):471-476.
95. Falagas ME, Mourtzoukou EG, Polemis M, Vatopoulos AC, Greek System for Surveillance of Antimicrobial Resistance. Trends in antimicrobial resistance of Acinetobacter baumannii clinical isolates from hospitalised patients in Greece and treatment implications. Clin Microbiol Infect 2007 Aug;13(8):816-819.
96. Roumbelaki M, Kritsotakis EI, Tsioutis C, Tzilepi P, Gikas A. Surveillance of surgical site infections at a tertiary care hospital in Greece: incidence, risk factors, microbiology, and impact. Am J Infect Control 2008 Dec;36(10):732-738.
97. Vatopoulos A. High rates of metallo-beta-lactamase-producing Klebsiella pneumoniae in Greece – a review of the current evidence. Euro Surveill 2008 Jan 24;13(4):8023.
98. Giakoupi P, Maltezou H, Polemis M, Pappa O, Saroglou G, Vatopoulos A, et al. KPC-2- producing Klebsiella pneumoniae infections in Greek hospitals are mainly due to a hyperepidemic clone. Euro Surveill 2009 May 28;14(21):19218.
99. Lanzas C, Ayscue P, Ivanek R, Grohn YT. Model or meal? Farm animal populations as models for infectious diseases of humans. Nat Rev Microbiol 2010 Feb;8(2):139-148.
100. Wray C, Gnanou JC. Antibiotic resistance monitoring in bacteria of animal origin: analysis of national monitoring programmes. Int J Antimicrob Agents 2000 May;14(4):291-294.
101. Martel JL, Tardy F, Sanders P, Boisseau J. New trends in regulatory rules and surveillance of antimicrobial resistance in bacteria of animal origin. Vet Res 2001 MayAug;32(3-4):381-392.
102. Weir M, Rajic A, Dutil L, Uhland C, Bruneau N. Zoonotic bacteria and antimicrobial resistance in aquaculture: opportunities for surveillance in Canada. Can Vet J 2012 Jun;53(6):619-622.
103. Joint Expert Advisory Committee on Antibiotic Resistance (JETACAR). The use of antibiotics in food-producing animals: antibiotic-resistant bacteria in animals and humans. 1999.
104. Ministère de l’Agriculture, des Pêcheries et de l’Alimentation du Québec (MAPAQ). Surveillance de l’antibiorésistance. Rapport annuel, 2011. 2012.
105. de Jong A, Thomas V, Klein U, Marion H, Moyaert H, Simjee S, et al. Pan-European resistance monitoring programmes encompassing food-borne bacteria and target pathogens of food-producing and companion animals. Int J Antimicrob Agents 2013 May;41(5):403-409.
106. Fedorka-Cray P, Englen MD, Gray JT, Hudson C, Headrick ML. Programs for monitoring antimicrobial resistance. Anim Biotechnol 2002 07/01; 2014/05;13(1):43-54.
107. Silley P, de Jong A, Simjee S, Thomas V. Harmonisation of resistance monitoring programmes in veterinary medicine: an urgent need in the EU? Int J Antimicrob Agents 2011 Jun;37(6):504-512.
108. European Food Safety Authority, Working Group on Developing Harmonised Schemes for Monitoring Antimicrobial Resistance in Zoonotic Agents. Harmonised monitoring of antimicrobial resistance in Salmonella and Campylobacter isolates from food animals in the European Union. Clin Microbiol Infect 2008 Jun;14(6):522-533.
109. Schwarz S, Alesik E, Grobbel M, Lubke-Becker A, Wallmann J, Werckenthin C, et al. The BfT-GermVet monitoring program – aims and basics. Berl Munch Tierarztl Wochenschr 2007 Sep-Oct;120(9-10):357-362.
110. ITAVARM. Italian Veterinary Antimicrobial Resistance Monitoring. First Report. 2003.
111. Ishihara K, Kira T, Ogikubo K, Morioka A, Kojima A, Kijima-Tanaka M, et al. Antimicrobial susceptibilities of Campylobacter isolated from food-producing animals on farms (1999-2001): results from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Int J Antimicrob Agents 2004 Sep;24(3):261-267.
112. Harada K, Asai T. Role of antimicrobial selective pressure and secondary factors on antimicrobial resistance prevalence in Escherichia coli from food-producing animals in Japan. J Biomed Biotechnol 2010;2010:180682.
113. Kojima A, Ishii Y, Ishihara K, Esaki H, Asai T, Oda C, et al. Extended-spectrum-betalactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob Agents Chemother 2005 Aug;49(8):3533-3537.
114. Asai T, Kojima A, Harada K, Ishihara K, Takahashi T, Tamura Y. Correlation between the usage volume of veterinary therapeutic antimicrobials and resistance in Escherichia coli isolated from the feces of food-producing animals in Japan. Jpn J Infect Dis 2005 Dec;58(6):369-372.
115. Morley PS, Apley MD, Besser TE, Burney DP, Fedorka-Cray PJ, Papich MG, et al. Antimicrobial drug use in veterinary medicine. J Vet Intern Med 2005 JulAug;19(4):617-629.
116. McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin Infect Dis 2002 Jun 1;34 Suppl 3:S93-S106.
117. DeVincent SJ, Viola C. Introduction to animal antimicrobial use data collection in the United States: methodological options. Prev Vet Med 2006 Feb 24;73(2-3):105-109.
118. Grave K, Greko C, Kvaale MK, Torren-Edo J, Mackay D, Muller A, et al. Sales of veterinary antibacterial agents in nine European countries during 2005-09: trends and patterns. J Antimicrob Chemother 2012 Dec;67(12):3001-3008.
119. Viola C, DeVincent SJ. Overview of issues pertaining to the manufacture, distribution, and use of antimicrobials in animals and other information relevant to animal antimicrobial use data collection in the United States. Prev Vet Med 2006 Feb 24;73(2- 3):111-131.
120. Weese JS. Investigation of antimicrobial use and the impact of antimicrobial use guidelines in a small animal veterinary teaching hospital: 1995-2004. J Am Vet Med Assoc 2006 Feb 15;228(4):553-558.
121. Moon CS, Berke O, Avery BP, McEwen SA, Reid-Smith RJ, Scott L, et al. Rates and determinants of antimicrobial use, including extra-label, on Ontario sheep farms. Can J Vet Res 2011 Jan;75(1):1-10.
122. Rajic A, Reid-Smith R, Deckert AE, Dewey CE, McEwen SA. Reported antibiotic use in 90 swine farms in Alberta. Can Vet J 2006 May;47(5):446-452.
123. McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 2013 Jul;57(7):3348-3357.
124. Bax R, Bywater R, Cornaglia G, Goossens H, Hunter P, Isham V, et al. Surveillance of antimicrobial resistance–what, how and whither? Clin Microbiol Infect 2001 Jun;7(6):316-325.
125. Colombo AL, Janini M, Salomão R, Medeiros EA, Wey SB, Pignatari AC. Surveillance programs for detection and characterization of emergent pathogens and antimicrobial resistance: results from the Division of Infectious Diseases, UNIFESP. An Acad Bras Cienc 2009 Sep;81(3):571-587.
126. Stephen C, Parmley J, Dawson-Coates J, Fraser E, Conly J. Obstacles to developing a multinational report card on antimicrobial resistance for Canada: an evidence-based review. Microb Drug Resist 2007 Winter;13(4):251-260.
127. U.S. Centers for Disease Control. (2001). Updated Guidelines for evaluating public health surveillance systems: recommendations from the guidelines working group. MMWR 50: 1 – 35.
128. Wegener HC. Antibiotic resistance — Linking human and animal health. In: Institute of Medicine (U.S.). Improving Food Safety Through a One Health Approach: Workshop Summary. Washington (DC): National Academies Press (U.S.); 2012. A15. Available from: http://www.ncbi.nlm.nih.gov/books/NBK114485
APPENDICES
A. Systematic Search Protocol
B. References Identified in the Systematic Search Protocol
C. Survey Questionnaire
D. Survey Results
E. Recommendations of Previous Canadian AMR Conferences and Proceedings
F. Categorization of antimicrobial drugs based on importance in human medicine (CIPARS, 2008)
Appendix A.
Systematic Search Protocol
The following description provides details of the systematic search protocol that was developed to review available literature related to Canadian and international examples of AMR and AMU surveillance programs.
The principal aim of this literature review was to identify and describe the strengths and weaknesses of the major AMR surveillance and antibiotic utilization monitoring programs in Canada, concentrating on antibacterial agents used in human patients and major food animals (poultry, swine and cattle) and to provide recommendations for improving these programs. A systematic search protocol, through collaboration with a professional medical research librarian, was developed to assemble key information related to AMR and antibiotic use surveillance in Canada and to provide examples for comparative purposes from existing programs from around the world.
Specific objectives of the review
- To determine the core elements, including organisms surveyed, data collected and methodological differences, of antimicrobial resistance (AMR) surveillance programs in Canada and around the world (national and international review of existing programs);
- To summarize current national, provincial, and regional AMR surveillance programs/initiatives within Canada;
- To summarize current national, provincial, regional, and private antimicrobial use monitoring programs/initiatives in Canada;
- To identify gaps in the areas of both antimicrobial use and resistance surveillance based on existing international model systems; and
- To recommend strategies for optimizing antimicrobial use monitoring as well as AMR surveillance programs.
Clarification of the research questions and the scope of the review
The antibacterial agents for human and veterinary uses and agri-food are included in the report. Evidence was gathered to answer the following questions:
- What exists for surveillance of antibiotic-resistant organisms in Canada, federally, provincially/territorially, and locally?
- What exists for surveillance of antimicrobial usage in Canada, federally, provincially territorially, regionally, institutionally, and locally?
- For the above surveillance systems, what information is gathered? To who is the data reported? How quickly is it reported?
- What international models exist for the collection, reporting and use of the data in monitoring resistance and guiding utilization practice?
- What provincial/national/international models for legislation and restrictions of use of antimicrobials exist?
- What surveillance systems for both usage and AMR surveillance have been tried in Canada, what has worked, what has failed and why has it worked/failed?
Inclusion and exclusion criteria
Inclusion criteria:
Agent:
• Only studies and surveillance systems focusing on antibacterial agents were considered
Types of studies:
• Peer-reviewed studies, which are broadly defined to include both quantitative and qualitative investigations, reviews, position papers, and guidelines on antibacterial agents only.
• Credible “grey literature” (e.g. technical reports from government agencies or scientific research groups, working papers from research groups or committees, research conference presentations, etc.)
• Surveillance reports
Study population:
• Human surveillance programs
i. Antibacterial drug utilization
ii. Antibacterial drug resistance
• Veterinary surveillance programs (Poultry (including turkeys), swine, cattle, fish)
i. Antibacterial drug utilization
ii. Antibacterial drug resistance
Types of interventions:
• Studies and surveillance systems examining prescription of antibiotics
• Stewardship/monitoring the use of antibiotics
• AMR surveillance programs
• AMR prevention and control programs/policies
• Studies on growth promotion in animals
• Studies related to zoonotic transfer of resistant microorganisms to/from pets
• Consequences of AMR in the environment
Outcomes:
• Accurate representation of utilization patterns and resistance profiles of antibacterials
• Description of the effectiveness (evaluation) of the existing antibiotic use and antibiotic resistance surveillance programs
• Effective policies
Time and place:
• Literature dating from 1990 to May 2013
• Historical papers dating back to 1970 (or earlier in specific contexts) was included based on references from the papers that met the inclusion criteria
Initial searches focused on studies conducted in Canada, Europe, the United States, Australia, New Zealand, Japan, Brazil, India, China, Russia, Israel and South Africa.
Exclusion criteria:
• Popular press, news reports
• Literature mentioning zoonosis but not focused on AMR
• Literature that did not focus on antimicrobial resistance or utilization
• Literature that focused on antiviral use and antiviral resistance
• Literature focusing on antifungal use and antifungal resistance
• Literature focusing on disinfectant use and disinfectant resistance
• Literature focusing on microbial resistance to metals (including mercury, trace metal resistance on plasmids, etc.)
• Literature focusing on topical antimicrobials, such as mupirocin, gramicidin, polymyxin, bacitracin, fucidin, sulfamylon, silver sulfadiazine, and AG sulfadiazine-CHG, etc.
• Literature concerning plant agriculture
• Literature concerning animals not mentioned in the inclusion criteria
Literature Search Strategy
In order to adequately address the six research questions in this systematic review, multiple approaches were employed. Additional information was secured through grey literature searches and by contacting relevant organizations and government ministries. The names of key authors and published papers provided by the research team supported the development of the search strategy both for the published and grey literature. The core committee the systematic search protocol prior to execution to ensure all relevant terms had been included. Multiple searches were conducted as an iterative process as new information was revealed based on the information gathered from previous searches. Documentation was maintained such that all searches are reproducible for the purposes of transparency.
Published Literature
The three key concepts for searching the bibliography databases focused on surveillance, drug utilization and drugs/infections (included drug resistance and antimicrobial cross resistance). Drug resistance was also combined with drugs/infections. A preliminary list of anti-infective agents and infections was compiled. Preliminary search models were constructed (Figures 2.1 and 2.2) with the search initially limited to Canada. Preliminary searches combining the concepts indicated that the number of studies was quite small for all combinations of the concepts. Subsequent searches removed the Canadian studies and initially limited to reviews and then focused the search on aspects of the research questions that were not covered or were insufficiently covered through reviews. The concepts for ‘veterinary surveillance’ and ‘drug use’ were similar but focused on animal diseases and veterinary drugs. The concept described by the term ‘animals’ included food animals and companion animals. The concept ‘animals’ when combined with drug utilization or drug resistance also indicated relevant papers when preliminary searches were performed.

Figure 1: Preliminary search model for human surveillance/drug utilization.
* Drug resistance was also combined as: Drug resistance AND (anti-infective agents OR infection OR antimicrobial resistance OR colonization)
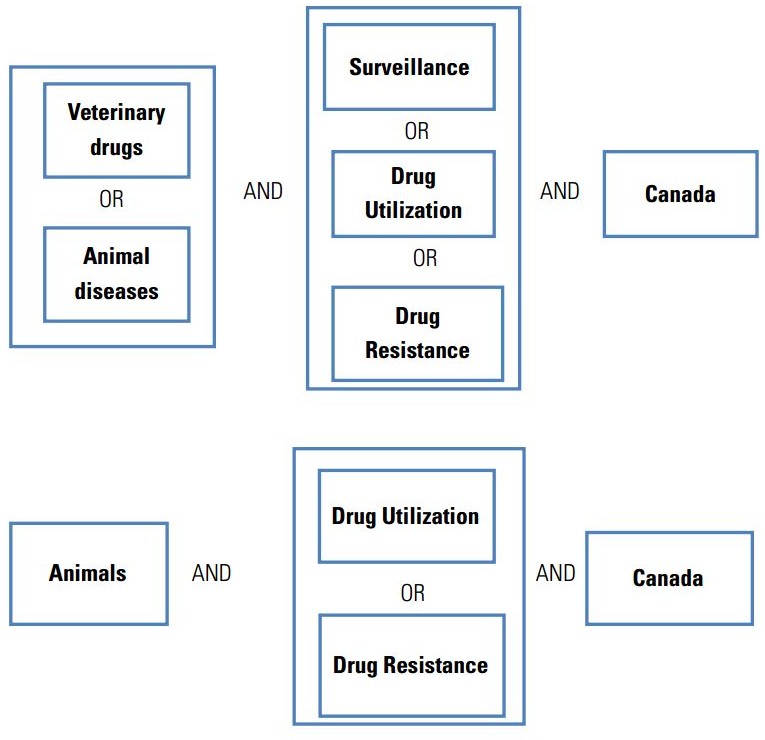
Figure 2: Preliminary search model for veterinary surveillance/drug utilization
Both subject headings and keywords were included in the searches. A template search was created in MEDLINE (OvidSP), which also incorporated relevant terms as keywords from other sources. This template search was translated into other databases using the appropriate syntax. There was no language restrictions placed on search results. When the searches were completed, the team decided whether complete translation was required for papers not in English, French, Spanish and Chinese that met the inclusion criteria. The searches were not limited to specific countries (other than the initial Canadian searches) as this would have reduced the sensitivity of the searches. Searches focused on the time period of 1990 to the present (2012) as most studies prior to the 1990s may not be applicable. Important historical papers back to the 1970s were examined once the main search was completed.
The key subject areas that the review encompassed included biology, medicine, nursing, pharmacy, public health, veterinary medicine and environmental health. These subjects cover a variety of databases which are listed below. The review was not limited to these databases. Monographs were also searched for comprehensive and historical information relating to the research questions. Subject terms were used to search the key concepts in the library databases. Initial searches focused on libraries at the national level which, in some cases, led to others searches on specific authors or titles.
To increase the sensitivity of database searches other approaches were used to capture missing papers from the published literature. The research team provided a list of authors who are experts in the subject areas pertinent to this review. These names were searched in the appropriate databases and their CVs for any missing papers. The references from papers that met the inclusion criteria were also reviewed to capture missing papers. All papers meeting the inclusion criteria were searched in the Web of Science, SCOPUS and MEDLINE for cited and related papers to ensure papers were not missed. New papers found through these methods went through the same selection process as those found in the database searches.
Grey Literature
Internet searches were the primary means for identifying applicable grey literature. Initial searches were conducted using Google but other search engines were also employed as some are better for languages other than English. A list of the main search engines was reviewed (http://www.thesearchenginelist.com/) and selected as needed.
The websites of key organizations were searched through the Internet for research articles, position papers and other relevant literature pertaining to the research questions. These sites were also reviewed for academic, government, community and other appropriate contacts. The associations related to the following subjects were searched:
- Microbiology
- Infectious diseases
- Public health
- Medical and dental
- Pharmaceutical
- Veterinary medicine
- Environmental health
- Agriculture (including provincial and regional)
- Dairy
- Meat and meat products
- Poultry and poultry products
- Fish and seafood
- Livestock
- Pet food and animal feed
Websites from Canadian government agencies at both the federal and provincial level were also searched as well as specific public health agencies. Key websites from government agencies and NGOs in specified countries were also included.
Conference proceedings were searched for papers, abstracts, and researchers using the key search concepts. These searches occurred through several databases including PapersFirst, ProceedingsFirst and the Web of Science. The websites of proceedings from selected associations were searched for relevant conference papers.
Professional Contacts
Key contacts through academic and professional affiliations and surveillance programs in Canada and other countries provided valuable information regarding research not accessible in the published literature, programs that are involved in surveillance and monitoring drug resistance and access to other contacts.
Study Selection
Papers were selected by means of a two-step process. Once a search had been completed and duplicates removed, titles and abstracts were reviewed by two reviewers independently. Their selection was based on broad inclusion criteria. The reviewers compared their selection and resolved any disagreements by discussion and when needed with a third party’s participation. The full text of selected papers was retrieved and the reviewers repeated the process when selecting the papers that met the inclusion criteria. Reviewers were not blinded to any components of the papers during their review.
Stage 1
Two reviewers independently reviewed titles and abstracts and made their selection based on 1) meeting the inclusion criteria 2) possibly meeting the inclusion criteria and were included for consideration 3) did not meet our inclusion criteria. 1 and 2 were automatically selected and articles from 3 are excluded. Selection was based on a broad view of the inclusion criteria as initially only titles and abstracts were read. Both reviewers made their selection, compared them and consulted with the principle investigators before final inclusion.
Stage 2
The process was repeated with the selected references from stage 1 but review of the full text of these papers was conducted by the two reviewers independently. Each paper had to meet critical aspects of the inclusion criteria by using a checklist of necessary components. Reasons for papers being excluded were recorded on the checklist. All papers not meeting the inclusion criteria were kept and archived with project records. Reviewers were not blind to authors in either stage in order to reduce selection bias.
Data Extraction Strategy
Two reviewers independently extracted the data using a pre-designed data extraction form. The data extraction form was pilot tested with several studies by the two reviewers to ensure all critical data was extracted and that decision rules and coding are completed in a similar manner. Disagreements were resolved by discussion with consultation with a third party in the case of further disagreement. Data from studies with multiple publications was extracted and reported as a single study.
The following data were extracted:
• Details of the study population and baseline characteristics of research groups
• Details of the care setting (e.g., physicians, dentists, nurses, midwives, veterinarians, pharmacists)
• Objective of the study
• Study methodology
• Study outcomes
• Specific information concerning antimicrobial resistance.
Quality Assessment Strategy
Two reviewers independently used a structured form and established process to undertake quality assessment. Final selected papers were reviewed using the Grading of Recommendations Assessment, Development and Evaluation (GRADE) system for assessment of quality of evidence. For the purposes of this review, the GRADE system classified the quality of evidence in one of three levels—high, moderate, and low (the very low classification level that is typically included in the GRADE system was combined into the ‘low’ level.
Disagreements were resolved by discussion and with consultation with third party in the case of remaining disagreement. The information on quality assessment was presented in table form and summarised within the text of the reports. The quality of studies were assessed using a validated checklist appropriate to the study type (i.e. quantitative versus qualitative).
Methods of analysis/synthesis
Initial literature analysis involved creation of a series of surveillance program comparison tables. Separate comparison tables for surveillance of antibiotic use, antimicrobial resistance in both human and veterinary medicine were prepared. These comparison tables were categorized as Canadian or international surveillance programs and studies. Data from these preliminary comparisons was then synthesized into the narrative summaries included in the formal literature search. The narrative summary of the studies included an explanation of the characteristics and findings of each of the included studies and the results for each of the specified outcome measures that were abstracted from the publications.
Documentation
The review process was tracked through a number of methods to provide thorough documentation. MS Word was used to record all search strategies through databases and the Internet. Unpublished documents and citations from database searches were downloaded or manually entered into RefWorks. Counts from citation results were recorded and dated and tracked throughout the identification of research papers. The research process was documented at each step to demonstrate the rigour of the review and to ensure study replication or updating if required. The PRISMA Statement (Figure 2.3) was used to report applicable aspects of this systematic review.
Results of the systematic literature search
A total of twenty databases were searched using the keywords and inclusion/exclusion criteria defined in the search protocol. Reference records were added electronically or manually into a RefWorks account for screening and evaluation. Duplicates were removed before all reference records were assembled. The electronic databases that were searched included:
1. MEDLINE
2. Cochran Database of Systematic Reviews (CDSR)
3. EMBASE (OvidSP)
4. Web of Science
5. BIOSIS Previews
6. CINAHL – Cumulated Index to Nursing and Allied Health Literature
7. TOXNET
8. LILACS
9. Informit (AGIS Plus Text)
10. CCOHS Web Information Service
11. JSTOR Collection
12. Environmental Sciences & Pollution Management
13. Pollution Abstracts
14. CAB Direct (CAB Abstracts & Global Health)
15. AGRICOLA
16. Canadian Periodical Index
17. Food Science and Technology Abstracts – FSTA
18. WAVES (Fisheries & Oceans Canada)
19. ASFA – Aquatic Sciences & Fisheries Abstracts
20. Google Scholar
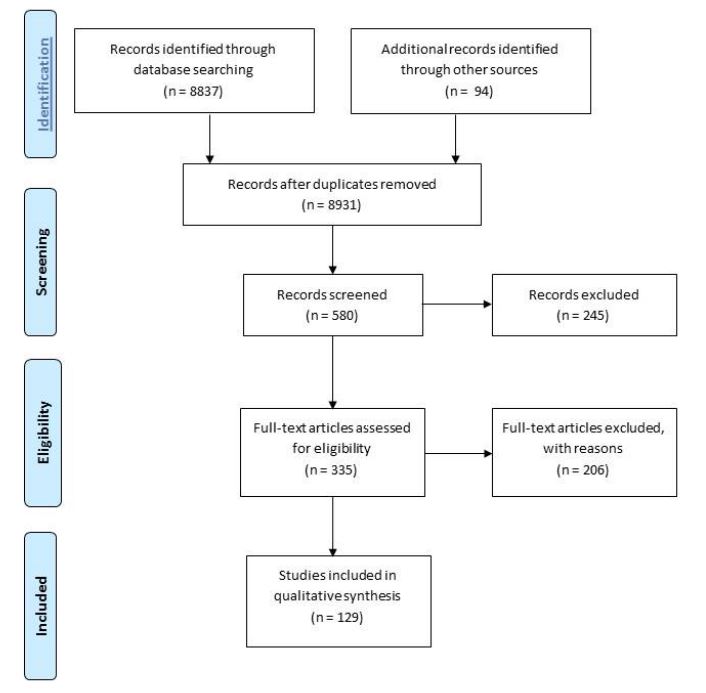
Figure 3: PRISMA 2009 Flow Diagram—AMU/AMR Surveillance
Primary Search Terms
Antibiotics: Drugs
| Antibiotics | Aminoglycoside | Amoxicillin |
| Amoxicillin trihydrate | Ampicillin (ampicillin sodium) | Arsphenamine |
| Azlocillin | Azithromycin | Aztreonam |
| Bacitracin | Beta-lactam | Carbapenems |
| Carbenicillin | Cefaclor | Cefadroxil |
| cefepime | Cefixime | Cefdinir |
| Cefditoren | Cefotaxime | Cefovecin |
| Cefpodoxime | Ceftazidime | Ceftibuten |
| Ceftriaxone | Cefepime | Ceftaroline fosamil |
| Ceftiofur | Ceftobiprole | Cefprozil |
| Cefuroxime | Cephalexin | Cephalosporin |
| Chloramphenicol | Ciprofloxacin | Clarithromycin |
| Clindamycin | Clotrimazole | Colistin |
| Daptomycin | Demeclocycline | Dicloxacillin |
| Doripenem | Doxycycline | Enoxacin |
| Enrofloxacin | Ertapenem | Florfenicol |
| Flucloxacillin | Fosfomycin | Furazolidone |
| Fusidic acid | Gatifloxacin | Gentamicin |
| Geldanamycin | Glycopeptide | Glycylcycline |
| Herbimycin | Imipenem/Cilastatin | Kanamycin |
| Levofloxacin | Lincomycin | Lincosamide |
| Linezolid | Lipopeptide | Lomefloxacin |
| Macrolide | Mafenide | Meropenem |
| Methicillin | Metronidazole | Minocycline |
| Monobactam | Moxifloxacin | Nalidixic acid |
| Neomycin | Netilmicin | Nitrofuran |
| Tetracycline (Chlortetracylcine; Oxytetracycline; Demeclocyline; Doxycycline; Lymecycline; Meclocycline; Methacycline; Minocycline; Rolitetracycline) | Thiamphenicol | Ticarcillin |
| Tigecycline | Tobramycin | Trimethoprim |
| Trimethoprim-Sulfamethoxazole (Cotrimoxazole) (TMP-SMX) | Vancomycin | Amoxicillin/clavulanate |
| Ampicillin/sulbactam | Piperacillin/tazobactam | Ticarcillin/clavulanate |
Surveillance
National program
Provincial program
Territorial program
Laboratory
Monitoring
Canada
United States
World
European countries
Use
Prescription
Human
Animal
Veterinary
Animal
Food animal
Farm
Companion animal
Pets
Wildlife
Zoonosis
Growth promotion
Antibiotic resistance: Antimicrobial resistance
Humans
Animals
Agronomy
Agriculture
Environment
Human health
Hospital case
Community case
Health care
Animal health
Hospital case
Community case
Infection/pathogen
Campylobacter
Carbapenem-resistant enterobacteriaceae (CRE)
VIM
NDM-1
KPC
Clostridium difficile (CDAD)
Enterococci
Extended-spectrum B-lactamase (ESBL)
Methicillin-resistant Staphylococcus aureus (MRSA)
Salmonella
Streptococcus pneumoniae
Vancomycin intermediate Staphylococcus aureus (VISA)
Vancomycin-resistant enterococci (VRE)
Vancomycin resistant Staphylococcus aureus (VRSA)
Appendix B.
References Identified in the Systematic Search Protocol
The following list contains the 129 references that met all or nearly all of the inclusion criteria and the quality criteria as defined in the systematic search protocol. Many, but not all, of these documents are referenced in the final report of the project.
1. Adam, HJ; Louie, L; Watt, C; Gravel, D; Bryce, E; Loeb, M; Matlow, A; McGeer, A; Mulvey, MR; Simor, AE. (2010). Detection and characterization of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates in Canada: Results from the Canadian Nosocomial Infection Surveillance Program, 1995-2006. Antimicrob. Agents Chemother. 54(2): 942–945.
2. Adam, HJ; Baxter, MR; Davidson, RJ; Rubinstein, E; Fanella, S; Karlowsky, JA; Lagacé-Wiens, PRS; Hoban, DJ; Zhanel, GG. (2013). Comparison of pathogens and their antibiotic resistance patterns in paediatric, adult and elderly patients in Canadian hospitals. J. Antimicrob. Chemother. 68 (Suppl 1): i31–i37.
3. Adriaenssens, N; Coenen, S; Versporten, A; Muller, A; Minalu, G; Faes, C; Vankerckhoven, V; Aerts, M; Hens, N; Molenberghs, G; Goossens, H. European Surveillance of Antimicrobial Consumption (ESAC): Outpatient antibiotic use in Europe (1997–2009). (2011a). J. Antimicrob. Chemother. 66 (suppl 6): vi3–vi12.
4. Adriaenssens, N; Coenen, S; Versporten, A; Muller, A; Minalu, C; Faes, C; Vankerckhoven, V; Aerts, M; Hens, N; Molenberghs, G; Goossens, H. (2011b). European Surveillance of Antimicrobial Consumption (ESAC): Outpatient macrolide, lincosamide and streptogramin (MLS) use in Europe (1997–2009). J. Antimicrob. Chemother. 66(Suppl 6): vi37–vi45.
5. Adriaenssens, N; Coenen, S; Versporten, A; Muller, A; Minalu, C; Faes, C; Vankerckhoven, V; Aerts, M; Hens, N; Molenberghs, G; Goossens, H. (2011c). European Surveillance of Antimicrobial Consumption (ESAC): Outpatient quinolone use in Europe (1997–2009). J. Antimicrob. Chemother. 66(Suppl 6): vi47–v56.
6. Adriaenssens, N; Coenen, S; Versporten, A; Muller, A; Vankerckhoven, V; Goossens, H. (2011d). European Surveillance of Antimicrobial Consumption (ESAC): Quality appraisal of antibiotic use in Europe. J. Antimicrob. Chemother. 66(Suppl 6): vi71–vi77.
7. Andrade, S; Sader, HS; Barth, A; Ribeiro, J; Zoccoli, C; Pignatari, AC; Gales, AC. (2008). Antimicrobial susceptibility of Gram-negative bacilli isolated in Brazilian hospitals participating in the SENTRY Program (2003-2008). Braz. J. Infect. Dis. 12(Suppl. 2): 3–9.
8. Apisarnthanarak, A; Buppunharun, W; Tiengrim, S; Sawanpanyalert, P; Aswapokee, N. (2009). An overview of antimicrobial susceptibility patterns for gram-negative bacteria from the national antimicrobial resistance surveillance Thailand (NARST) program from 2000 to 2005. J. Med. Assoc. Thai. 92(Suppl 4): S91-S94.
9. Bager, F. (2000). DANMAP: Monitoring antimicrobial resistance in Denmark. Int. J. Antimicrob. Agents. 14: 271-274. 10. Ballow, CH; Jones, RN; Biedenbach, DJ; North American ZAPS Research Group. (2002a). A multicenter evaluation of linezolid antimicrobial activity in North America. Diagn. Microbiol. Infect. Dis. 43(1): 75–83.
11. Ballow, CH; Biedenbach, DJ; Rossi, F; Jones, RN; LA-ZAPS Study Group. (2002b). Multicenter assessment of the linezolid spectrum and activity using the disk diffusion and E-test methods: Report of the Zyvox(R) antimicrobial potency study in Latin America (LA-ZAPS). Braz. J. Infect. Dis. 6(3): 100–109.
12. Bax, R; Bywater, R; Cornaglia, G; et al. 2001. Surveillance of antimicrobial resistance – What, how and wither? Clin Microbiol Infect. 7: 316–325.
13. Bell, JM; Turnidge, JD; Jones, RN; The SENTRY Asia-Pacific Participants. (2003). Prevalence of extended-spectrum ß-lactamase-producing Enterobacter cloacae in the Asia-Pacific region: Results from the SENTRY Antimicrobial Surveillance Program, 1998 to 2001. Antimicrob. Agents Chemother. 47: 3989– 3993.
14. Bergman, M; Huikko, S; Pihlajamäki, M; Laippala, P; Palva, E; Huovinen, P; Seppälä, H. (2004). Effect of macrolide consumption on erythromycin resistance in Streptococcus pyogenes in Finland in 1997–2001. Clin. Infect. Dis. 38: 1251–1256.
15. Bergman, M; Nyberg, S; Huovinen, P; Paakkari, P; Hakanen; The Finnish Study Group for Antimicrobial Resistance. (2009). Association between antimicrobial consumption and resistance in Escherichia coli. Antimicrob. Agents Chemother. 53(3): 912–917.
16. Biedenbach, DJ; Bell, JM; Sader, HS; Fritsche, TR; Jones, RN; Turnidge, JD. (2007). Antimicrobial susceptibility of Gram-positive bacterial isolates from the Asia-Pacific region and an in vitro evaluation of the bactericidal activity of daptomycin, vancomycin and teicoplanin: A SENTRY Program report (2003- 2004). Int. J. Antimicrob. Agents 30: 143–149. 17. Castanheira, M; Deshpande, LM; Mathai, D; Bell, JM; Jones, RN; Mendes, RE. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: Report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob. Agents Chemother. 55: 1274–1278.
18. Coenen, S; Ferech, M; Dvorakova, K; Hendrick, E; Suetens, C, Goossens, H. (2006a). European surveillance of antimicrobial consumption (ESAC): Outpatient cephalosporin use in Europe. J. Antimicrob. Chemother. 58(2): 413–417.
19. Coenen, S; Ferech, M; Malhotra-Kumar, S; Hendricks, E; Suetens, C; Goossens, H. (2006b). European Surveillance of Antimicrobial Consumption (ESAC): Outpatient macrolide, lincosamide and streptogramin (MLS) use in Europe. J. Antimicrob. Chemother. 58: 418–422.
20. Coenen, S; Adriaenssens, N; Versporten, A; Muller, A; Minalu, C; Faes, C; Vankerckhoven, V; Aerts, M; Hens, N; Molenberghs, G; Goossens, H. (2011). European Surveillance of Antimicrobial Consumption (ESAC): Outpatient use of tetracyclines, sulphonamides and trimethoprim, and other antibacterials in Europe (1997–2009). J. Antimicrob. Chemother. 66(Suppl 6): vi57–vi70.
21. Colombo, AL; Janini, M; Salomão, R; Medeiros, EA; Wey, SB; Pignatari, AC. (2009). Surveillance programs for detection and characterization of emergent pathogens and antimicrobial resistance: Results from the division of infectious diseases, UNIFESP. An Acad Bras Cienc. 81(3): 571-587.
22. Conly, JM; Ofner-Agostini, M; Paton, S; Johnston, L; Mulvey, MR; Kureishi, A; Nicolle, L; Matlow, A. (2001), et al. The emerging epidemiology of VRE in Canada: Results of the CNISP Passive Reporting Network, 1994 to 1998. Can. J. Infect. Dis. 12(6): 364–370.
23. Conly, JM. (2002). Antimicrobial resistance in Canada. Can. Med. Assoc. J. 167(8): 885-891.
24. Conly, JM; McEwen, S; Hutchinson, J; Boyd, N; Callery, S; Bryce, E. (2004). Canadian Committee on Antibiotic Resistance report. Can. J. Infect. Dis. Med. Microbiol. 15(5): 257–260.
25. Cornaglia, G; Hryniewicz, W; Jarlier, V; Kahlmeter, G; Mittermayer, H; Stratchounski, L; Baquero, F. (2004). European recommendations for antimicrobial resistance surveillance. Clin. Microbiol. Infect. 10(4): 349–383.
26. Dejsirilert, S; Tiengrims, S; Sawanpanyalert, P; Aswapokee, N; Malathum, K. (2009). Antimicrobial resistance of Acinetobacter baumannii: Six years of National Antimicrobial Resistance Surveillance Thailand (NARST) surveillance. J. Med. Assoc. Thai. 92 (Suppl 4): S34-S45.
27. Dejsirilert, S; Tienkrim, S; Ubonyaem, N; Sawanpanyalert, P; Aswapokee, N; Suankratay, C. (2009). National antimicrobial resistance surveillance among clinical isolates of Streptococcus pneumoniae in Thailand. J. Med. Assoc. Thai. 92 (Suppl 4): S19-S33.
28. de Sande-Bruinsma, NV; Grundmann, H; Verloo, D; Tiemersma, E; Monen, J; Goossens, H; Ferech, M. (2008). Antimicrobial drug use and resistance in Europe. Emerg. Infect. Dis. 14: 1722–1730. 29. Deshpande, LM; Fritsche, TR; Jones, RN. (2004). Molecular epidemiology of selected multidrug-resistant bacteria: A global report from the SENTRY Antimicrobial Surveillance Program. Diagn. Microbiol. Infect. Dis. 49: 231–236. 30. Deshpande, LM; Jones, RN; Fritsche, TR; Sader, HS. (2006). Occurrence and characterization of carbapenamase-producing Enterobacteriaceae: Report from the SENTRY Antimicrobial Surveillance Program (2000-2004). Microb. Drug. Resist. 12: 223–230. 31. Doern, GV; Pfaller, MA; Erwin, ME; Brueggemann, AB; Jones, RN. (1998). The prevalence of fluoroquinolone resistance among clinically significant respiratory tract isolates of Streptococcus pneumoniae in the United States and Canada – 1997 results from the SENTRY Antimicrobial Surveillance Program. Diagn. Microbiol. Infect. Dis. 32: 313–316. 32. De Jong, A; Thomas, V; Klein, U, Marion, H; Moyaert, H; Simjee, S; Vallé. (2013). Pan-European resistance monitoring programmes encompassing food-borne bacteria and target pathogens of food-producing and companion animals. Int. J. Antimicrob. Agents. 41(5): 403–409.
33. Drewe, JA; Hoinville, LJ; Cook, AJC; Floyd, T; Stärk, KDC (2012). Evaluation of animal and public health surveillance systems: systematic review. Epidemiology and Infection. 140: 575-590.
34. European Centre for Disease Prevention and Control (ECDC). (2012). EARS-Net Reporting Protocol. Version 2. Available at: http://www.ecdc.europa.eu/en/activities/surveillance/EARSNet/Documents/2010_EARS-Net_Reporting%20Protocol.pdf. Accessed: May 18, 2013.
35. European Food Safety Authority. (2008). Harmonised monitoring of antimicrobial resistance in Salmonella and Campylobacter isolates from food animals in the European Union. Clin. Microbiol. Infect. 14: 522–533.
36. Faes, C; Molenberghs, G; Hens, N; Muller, A; Goossens, H; Coenen, S. (2011). Analysing the composition of outpatient antibiotic use: A tutorial on compositional data analysis. J. Antimicrob. Chemother. 66(Suppl 6): vi89–vi94. 37. Farrell, DJ; Castanheira, M; Mendes. RE; Sader, HS; Jones, RN. (2012). In vitro activity of ceftaroline against multidrug-resistant Staphylococcus aureus and Streptococcus pneumoniae: A review of published studies and the AWARE Surveillance Program (2008-2010). Clin. Infect. Dis. 55(Suppl 3): S206–S214.
38. Felmingham, D; White, AR; Jacobs, MR; Appelbaum, PC; Poupard, J; Miller, LA; Grüneberg, RN. (2005). The Alexander Project: The benefits from a decade of surveillance. J. Antimicrob. Chemother. 56 (Suppl S2): 3–21.
39. Ferech, M; Coenen, S; Malhotra-Kumar, S; Dvorakova, K; Hendrickx, E; Suetens, C; Goossens, H. (2006a). European Surveillance of Antimicrobial Consumption (ESAC): Outpatient antibiotic use in Europe. J Antimicrob. Chemother. 58: 401– 407.
40. Ferech, M; Coenen, S; Dvorakova, K; Hendrickx, E; Suetens, C; Goossens, H. (2006b). European Surveillance of Antimicrobial Consumption (ESAC): Outpatient penicillin use in Europe. J. Antimicrob. Chemother. 58(2): 408–412.
41. Ferech, M; Coenen, S; Malhotra-Kumar, S; Dvorakova, K; Hendrickx, E; Suetens, C; Goossens, H. (2006). European Surveillance of Antimicrobial Consumption (ESAC): Outpatient quinolone use in Europe. J. Antimicrob. Chemother. 58: 423– 427.
42. Finch, R. (2002). Antibiotic resistance – from pathogen to disease surveillance. Clin. Microbiol. Infect. 8(6): 317–320.
43. Fluit, AC; van der Bruggen, JT; Aarestrup, FM; Verhoef, J; Jansen, WT. (2006). Priorities for antibiotic resistance surveillance in Europe. Clin. Microbiol. Infect. 12(5): 410–417.
44. Forrester, L; Collet, JC; Mitchell, R; Pelude; L; Henderson, E; Vayalumkal, J; Leduc, S; Ghahreman, S; Weir, C;, Gravel, D. (2012). How reliable are national surveillance data? Findings from an audit of Canadian methicillin-resistant Staphylococcus aureus surveillance data. Am. J. Infect. Control. 40(2): 102–107
. 45. Frank, U, for the BURDEN study Group. (2007). The BURDEN project – assessing the burden of resistance and disease in Europe. Eurosurveillance 12(2): pii-3112.
46. Fridkin, SK; Steward, CD; Edwards, JR; Pryor, ER; McGowan, JE; Archibald, LK; Gaynes, RP; Tenover, FC. (1999). Surveillance of antimicrobial use and antimicrobial resistance in United States hospitals: Project ICARE phase 2. Project Intensive Care Antimicrobial Resistance Epidemiology (ICARE) hospitals. Clin. Infect. Dis. 29: 245–252.
47. Gales, AC; Castanheira, M; Jones, RN; Sader, HS. (2012). Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: Results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008-2010). Diagn. Microbiol. Infect. Dis. 73(4): 354–360.
48. Gilca, R; Fortin, E; Frenette, C; Longtin, Y; Gourdeau, M. (2012). Seasonal variations in Clostridium difficile infections are associated with influenza and respiratory syncytial virus activity independently of antibiotic prescriptions: A time series analysis in Quebec, Canada. Antimicrob. Agents Chemother. 56(2): 639–646.
49. Goossens, H; Grabein, B. (2005). Prevalence and antimicrobial susceptibility data for extended-spectrum beta-lactamase- and AmpC-producing Enterobacteriaceae from the MYSTIC Program in Europe and the United States (1997–2004). Diagn. Microbiol. Infect. Dis. 53: 257–264.
50. Goossens, H; Ferech, M; Vander Stichele, R; Elseviers, M. (2005). Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet 365: 579–587. 51. Goossens, H; Ferech, M; Coenen, S; Stephens, P. (2007). European surveillance of antimicrobial consumption project group. Comparison of outpatient systemic antibacterial use in 2004 in the United States and 27 European countries. Clin. Infect. Dis. 44: 1091–1095 52. Goossens, H. (2009). Antibiotic consumption and link to resistance. Clin. Microbiol. Infect. 15 (Suppl. 3): 12–15. 53. Grave, K; Greko, C; Kvaale, MK: Torren-Edo, J; Mackay, D; Muller, A; Moulin, G. (2012). Sales of veterinary antibacterial agents in nine European countries during 2005-09: Trends and patterns. J. Antimicrob. Chemother. 67(12): 3001– 3008.
54. Hoban, DJ; Biedenbach, DJ; Mutnick, AH; Jones, RN. (2003). Pathogen of occurrence and susceptibility patterns associated with pneumonia in hospitalized patients in North America: Results of the SENTRY Antimicrobial Surveillance Study (2000). Diagn. Microbiol. Infect. Dis. 45: 279–285.
55. Hoban, DJ; Zhanel, GG. (2013), Introduction to the CANWARD study (2007-11). J. Antimicrob. Chemother. 68 (Suppl 1): i3–i5.
56. Hutchinson, JM; Patrick, DM; Marra, F; Ng, H; Bowie, WR; Heule, L; Muscat, M; Monnet, D. (2004). Measurement of antibiotic consumption: A practical guide to the use of the anatomical therapeutic chemical classification and defined daily dose system methodology in Canada. Can. J. Infect. Dis. 15(1): 29–35.
57. Jenkins, SG; Farrell, DJ; Patel, M; Lavin, BS. (2005). Trends in anti-bacterial resistance among Streptococcus pneumoniae isolated in the USA, 2000–2003: PROTEKT US years 1–3. J. Infect. 51: 355–363. 58. Johnson, AP; Davies, J; Guy, R; Abernathy, J; Sheridan, E; Pearson, A; Duckworth, G. (2012). Mandatory surveillance of methicillin-resistant Staphylococcus aureus (MRSA) bacteraemia in England: The first 10 years. J. Antimicrob. Chemother. 67: 802–809.
59. Jones, ME; Jones, RN; Sader, H; Verhoef, J; Acar, J. (1998). Current susceptibilities of staphylococci to glycopeptides determined as part of an international resistance surveillance programme. SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 42: 119–121.
60. Jones, RN; Pfaller, MA. (2000). In vitro activity of newer fluoroquinolones for respiratory tract infections and emerging patterns of antimicrobial resistance: Data from the SENTRY antimicrobial surveillance program. Clin. Infect. Dis. 31(suppl 2): S16-S23.
61. Jones, RN; Ballow, CH; Biedenbach, DJ, ZAPS Study Group Medical Centers. (2001). Multi-laboratory assessment of the linezolid spectrum of activity using the Kirby-Bauer disk diffusion method: Report of the Zyvox Antimicrobial Potency Study (ZAPS) in the United States. Diagn. Microbiol. Infect. Dis. 40(1-2): 59–66.
62. Jones, RN; Mendes, C; Turner, PJ; Masterton, R. (2005). An overview of the Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) Program: 1997–2004. Diagn. Microbiol. Infect. Dis. 53: 247–256.
63. Jones, RN; Sader, HS; Moet, GJ; Farrell, DJ. (2010). Declining antimicrobial susceptibility of Streptococcus pneumoniae in the United States: Report from the SENTRY Antimicrobial Surveillance Program (1998-2009). Diagn. Microbiol. Infect. Dis. 68: 334–336.
64. Jones, RN; Sader, HS; Flamm, RK. (2013). Update of dalbavancin spectrum and potency in the USA: Report from the SENTRY Antimicrobial Surveillance Program (2011). Diagn. Microbiol. Infect. Dis. 75(3): 304–307. 65. Kaier, K; Wilson, C; Chaklye, M; Davey, PG; Suetens, C; Grundmann, H; de Kraker, M; Schumacher, M; Wolkewitz, M, Frank, U. (2008). Health and economic impacts of antibiotic resistance in European hospitals – Outlook on the BURDEN project. Infection 36(5): 492–494. 66. Koeth, LM; Miller, LA. (2005). Evolving concepts of pharmaceutical companysponsored surveillance studies. Clin. Infect. Dis. 41(Suppl 4): S279–S279.
67. Kojima, A; Ishihara, Y; Yamaguchi, K. (2005). Extended-spectrum-β-lactamaseproducing Escherichia coli strains isolated from farm animals from 1999 to 2002: Report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob. Agents Chemother. 49(8): 3533–3537.
68. Kuti, JL; Nicolau, DP. (2005). Making the most of surveillance studies: Summary of the OPTAMA program. Diagn. Microbiol. Infect. Dis. 53(4): 281-287.
69. Lagacé-Wiens, PRS; Adam, HJ; Low, DE; Blondeau, JM; Baxter, MR; Denisuik, AJ; Nichol, KA; Walkty, A, Karlowsky, JA; Mulvery, MR, Hoban, DJ; Zhanel, GG. (2013). Trends in antibiotic resistance over time among pathogens from Canadian hospitals: Results of the CANWARD study 2007-11. J. Antimicrob. Chemother. 68 (Suppl 1): i23–i29.
70. Lawton, RM; Fridkin, SK; Steward, CD; Edwards, JR; Pryor, ER; McGowan, JE; Archibald, LK; Gaynes, RP; Tenover, FC; Pichette, SC; Mohammed, J (Mohammed, J); Felicione, E; Hubert, S. (1999). Intensive Care Antimicrobial Resistance Epidemiology (ICARE) Surveillance Report, data summary from January 1996 through December 1997: A report from the National Nosocomial Infections Surveillance (NNIS) System. Am. J. Infect. Control. 27: 279–284. 71. Martel, JL; Tardy. F; Brisabois, A; Lailler, R; Coudert, M; Chaslus-Dancia, E. (2000). The French antibiotic resistance monitoring programs. Int. J. Antimicrob. Agents 14: 275-283.
72. Martel, JL; Tardy, F; Sanders, P; Boisseau, J. (2001). New trends in regulatory rules and surveillance of antimicrobial resistance in bacteria of animal origin. Vet. Res. 32(3-4): 381–392.
73. Martin, I; Jayaraman, G; Wong, T; Liu, G; Gilmour, M. (2011). Trends in antimicrobial resistance in Neisseria gonorrhoeae isolated in Canada: 2000-2009. Sex. Transm. Dis. 38(10): 892-895.
74. Masteron, R. (2008). The importance and future of antimicrobial surveillance studies. Clin. Infect. Dis. 47(Suppl 1): S21-S31.
75. Masterton, RG; Turner, PJ. (2006). Trends in antimicrobial susceptibility in UK centres: The MYSTIC programme (1997–2002). Int. J. Antimicrob. Agents. 27 (1): 69–72.
76. Masterton, RG; Kuti, JL; Turner, PJ; Nicolau, DP. (2005). The OPTAMA programme: Utilizing MYSTIC (2002) to predict critical pharmacodynamic target attainment against nosocomial pathogens in Europe. J. Antimicrob. Chemother. 55(1): 71-77.
77. McEwen, SA; Fedorka-Cray, PJ. (2002). Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34 (Suppl 3): S93–S106.
78. Minalu, G; Aerts, M; Coenen, S; Versporten, A; Muller, A; Adriaenssens, N; Beutels, P; Molenberghs, G; Goossens, H; Hens, N. (2011). Application of mixedeffects models to study the country-specific outpatient antibiotic use in Europe: A tutorial on longitudinal data analysis. J. Antimicrob. Chemother. 66(Suppl 6): vi79– vi87.
79. Mölstad, S; Cars, O; Struwe, J. (2008). Strama – A Swedish working model for containment of antibiotic resistance. Euro. Surveill. 13(46): 1–4.
80. Mootsikapun P, Trakulsomboon S, Sawanpanyalert P, Aswapokee N, Suankratay C. (2009). An overview of antimicrobial susceptibility patterns of gram-positive bacteria from National Antimicrobial Resistance Surveillance Thailand (NARST) program from 2000 to 2005. J. Med. Assoc. Thai. 92(Suppl 4):S87-S90.
81. Morfin-Otero, R; Rodriguez-Nkoriega, E; Desphande, LM; Sader, HS; Castanheira, M. (2009). Dissemination of a bla(VIM-2)-carrying integron among Enterobacteriaceae species in Mexico: Report from the SENTRY Antimicrobial Surveillance Program Microbial. Drug Resistance 15: 33–35.
82. Morris, AK; Masterton, RG. (2002). Antibiotic resistance surveillance: Action for international studies. J. Antimicrob. Chemother. 49(1): 7–10.
83. Morrow, BJ; Pillar, CM; Deane, J; Sahm, DF; Lynch, AS; Flamm, RK; Peterson, J; Davies, TA. (2013). Activities of carbapenem and comparator agents against contemporary US Pseudomonas aeruginosa isolates from the CAPITAL surveillance program. Diagn. Microbiol. Infect. Dis. 75(4): 412-416.
84. Neu, H C; Duma, R J; Jones, RN; McGowan, JE; O’Brien, TF; Sabath, LD et al. (1992). Antibiotic resistance. Epidemiology and therapeutics. Diagn. Microbiol. Infect. Dis. 15: 53S–60S.
85. Nissinen, A; Huovinen, P. (2000). FiRe works—the Finnish Study Group for Antimicrobial Resistance (FiRe). Euro. Surveill. 5: 133-135.
86. O’Brien, TF; Stelling, J. (2011). Integrated surveillance of the world’s infecting microbes and their resistance to antimicrobial agents. Clin. Microbiol. Rev. 24(2): 261-295.
87. PAHO (Pan American Health Organization. (2009). Informe Anual de la Red de Monitoreo / Vigilancia de la Resistencia a los Antibióticos. OPS/HDM/CD/A/541/09, Lima, Peru.
88. Pasteran, F; Albornoz, E; Faccone, D; Gomez, S; Valenzuela, C; Morales, M; Estrada, P; Valenzuela, L; Matheu, J; Guerriero, L; Arbizú, E; Calderón, Y; Ramon-Pardo, P; Corso, A. (2012). Emergence of NDM-1-producing Klebsiella pneumoniae in Guatemala. J. Antimicrob. Chemother. 67: 1795–1797.
89. Pfaller, MA; Jones, RN, Doern, GV; Kugler, K; The SENTRY Participants Group. (1998). Bacterial pathogens isolated from patients with bloodstream infections: Frequencies of occurrence and antimicrobial susceptibility patterns from the SENTRY antimicrobial surveillance program (United States and Canada, 1997). Antimicrob. Agents Chemother. 42(7): 1762–1770.
90. Pfaller, MA; Farrell, DJ; Sader, HS; Jones, RN. (2012). AWARE Ceftaroline Surveillance Program (2008-2010): Trends in resistance patterns among Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the United States. Clin. Infect. Dis. 55(Suppl 3): S187–S193.
91. PHAC (Public Health Agency of Canada). (2009). C-EnterNet Final Report – Canada’s National Integrated Enteric Pathogen Surveillance System. Ottawa, Ontario. Available at: http://198.103.98.45/c-enternet/pdf/sr-rs-2009-eng.pdf. Accessed Mar 2, 2013.
92. Poupard, J; Brown, J; Gagnon, R; Stanhope, MJ; Stewart, C. (2002). Methods for data mining from large multinational surveillance studies. Antimicrob. Agents Chemother. 46(8): 2409–2419.
93. Queenan, AM; Pillar, CM; Deane, J; Sahm, DF; Lynch, AS; Flamm, RK; Peterson, J; Davies, TA. (2012). Multidrug resistance among Acinetobacter spp. in the USA and activity profile of key agents: Results from CAPITAL Surveillance 2010. Diagn. Microbiol. Infect. Dis. 73: 267–270
94. Rempel, OR; Laupland, KB. (2009). Surveillance for antimicrobial resistant organisms: Potential sources and magnitude of bias. Epidemiol. Infect. 137: 1665– 1673.
95. Rhomberg PR, Deshpande LM, Kirby JT, Jones RN. (2007). Activity of meropenem as serine carbapenemases evolve in US medical centers: monitoring report from the MYSTIC Program (2006). Diagn. Microbiol. Infect. Dis. 59: 425– 432.
96. Rhomberg, PR; Jones, RN. (2009). Summary trend for the Meropenem Yearly Susceptibility Test Information Collection Program: A 10-year experience in the United States (1999–2008). Diagn. Microbiol. Infect. Dis. 65: 414–426.
97. Rossi, F; Baquero, F; Hsueh, PR; (2006). In vitro susceptibilities of aerobic and facultatively anaerobic Gram-negative bacilli isolated from patients with intraabdominal infections worldwide: 2004 results from SMART (Study for Monitoring Antimicrobial Resistance Trends). J. Antimicrob. Chemother. 58(1): 205–210.
98. Rybak, MJ. (2004). Increased bacterial resistance: PROTEKT US – an update. Ann. Pharmacother. 38(Suppl 9): S8-S13.
99. Sader, HS; Jones, RN; Gales, AC; Silva, JB; Pignatari, AC. (2004). SENTRY antimicrobial surveillance program report: Latin American and Brazilian results for 1997 through 2001. Braz. J. Infect. Dis. 8: 25–79.
100. Sader, H.S., Fritsche, T.R., Jones, R.N.. (2005). Potency and spectrum trends for cefepime tested against 65,746 clinical bacterial isolates collected in North American medical centers: Results from the SENTRY Antimicrobial Surveillance Program (1998 – 2003). Diagn. Microbiol. Infect. Dis. 52: 265–273.
101. Seppälä, H; Klaukka, T; Vuopio-Varkila, J; Muotiala, A; Helenius, H; Lager, K; Huovinen, P; The Finnish Study Group for Antimicrobial Resistance. (1997). The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A Streptococci in Finland. N Engl J Med. 337: 441–446.
102. Silley, P; de Jong, A; Simjee, S; Thomas, V. (2011). Harmonisation of resistance monitoring programmes in veterinary medicine: An urgent need in the EU? Int. J. Antimicrob. Agents. 37(6): 504–512.
103. Silley, P; Simjee, S; Schwarz, S. (2012). Surveillance and monitoring of antimicrobial resistance and antibiotic consumption in humans and animals. Rev. Sci. Tech. Off. Int. Epiz. 31(1): 105–129.
104. Starnino, S; Galarza, P; Carvallo, MET; Benzaken, AS; Ballesteros, AM; Sanabria, OM; Hernandez, AL; Carbajal, JLP; Borthagaray, G; Payares, D; Dillon, JR. (2012). Retrospective analysis of antimicrobial susceptibility trends (2000-2009) in Neisseria gonorrhoeae isolates from countries in Latin America and the Caribbean shows evolving resistance to ciprofloxacin, azithromycin and decreased susceptibility to ceftriaxone. Sex. Trans. Dis. 39(10): 813–821.
105. Stephen, C; Parmley, J; Dawson-Coates, J; Fraser, E; Conly, J. (2007). Obstacles to developing a multinational report card on antimicrobial resistance for Canada: An evidence-based review. Microb. Drug Resistance. 13(4): 251–259.
106. Stelling, JM; O’Brien, TF. (1997). Surveillance of antimicrobial resistance: The WHONET program. Clin. Infect. Disease. 24 (suppl 1): S157-S168.
107. Stelling, JM; Travers, K; Jones, RN; Turner, PJ; O’Brien, TF; Levy, SB. (2005). Integrating Escherichia coli antimicrobial susceptibility data from multiple surveillance programs. Emerg. Infect. Dis.11: 873–882.
108. Struwe, J. (2008). Fighting antibiotic resistance in Sweden – Past, present and future. Wien Klin Wochenschr. 120(9-10): 268–279.
109. Takahashi, T; Asai, T; Kojima, A; Harada, K; Ishihara, K; Morioka, A; Kijima, M; Tamura, Y. (2006). Present situation of national surveillance of antimicrobial resistance in bacteria isolated from farm animals in Japan and correspondence to the issue. Kansenshogaku Zasshi. 80(3): 185–195.
110. Toleman, MA; Simm, AM; Murphy, TA; Gales, AC; Biedenbach, DJ; Jones, RN; Walsh, TR. (2002). Molecular characterization of SPM-1: a novel metallobetalactamase isolated in Latin America: Report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50: 673–679.
111. Turner, PJ. (2000). MYSTIC (Meropenem yearly susceptibility test information collection): A global overview. J. Antimicrob. Chemother. 42 (Topic T2): 9–23.
112. Turner, PJ. (2005). Use of a program-specific website to disseminate surveillance data obtained from the MYSTIC (Meropenem Yearly Susceptibility Test Information Collection) Study. Diagn. Microbiol. Infect. Dis. 53: 273–279.
113. Turner, PJ. (2008). Meropenem activity against European isolates: Report on the MYSTIC (Meropenem Yearly Susceptibility Test Information Collection) 2006 results. Diagn. Microbiol. Infect. Dis. 60: 185–192.
114. United Nations. (2001). WHO Global Strategy for Containment of Antimicrobial Resistance. http://whqlibdoc.who.int/hq/2001/WHO_CDS_CSR_DRS_2001.2.pdf Accessed: January 12, 2013.
115. United States Centers for Disease Control (2013). Antibiotic Resistance Threats in the United States, 2013. U.S. Department of Health and Human Services Centers for Disease Control and Prevention. Atlanta Georgia, USA.
116. United States Centers for Disease Control. (2012). National Centre for Emerging and Zoonotic Infectious Diseases Strategic Plan 2012-2017. Atlanta, Georgia USA.
117. Vander Stichele, RH; Elseviers, MM; Ferech, M; Blot, S; Goossens, H. (2004). European Surveillance of Antimicrobial Consumption (ESAC): Data collection performance and methodological approach. Br. J. Clin. Pharmacol. 58: 419–428.
118. Versporten, A; Coenen, S; Adriaenssens, N; Muller, A; Minalu, G; Faes, C; Vankerckhoven, V; Aerts, M; Hens, N; Molenberghs, G, Goossens, H. (2011a). European Surveillance of Antimicrobial Consumption (ESAC): Outpatient penicillin use in Europe (1997–2009). J. Antimicrob. Chemother. 66(Suppl 6): vi13–vi23.
119. Versporten, A; Coenen, S; Adriaenssens, N; Muller, A; Minalu, G; Faes, C; Vankerckhoven, V; Aerts, M; Hens, N; Molenberghs, G, Goossens, H. (2011b). European Surveillance of Antimicrobial Consumption (ESAC): Outpatient cephalosporin use in Europe (1997–2009). J. Antimicrob. Chemother. 66(Suppl 6): vi25–vi35.
120. Viola, C; DeVincent, SJ. (2006). Overview of issues pertaining to the manufacture, distribution, and use of antimicrobials in animals and other information relevant to animal antimicrobial use data collection in the United States. Prev. Vet. Med. 73: 111–131.
121. Walter, R. (2005). Antimicrobial resistance: An update from the Canadian Committee on Antibiotic Resistance. Can. J. Infect. Dis. Med. Microbiol. 16(5): 309–311.
122. Wang, EEL, Einarson, TR; Kellner, JD; Conly, JM. (1999). Antibiotic prescribing for Canadian preschool children: Evidence of overprescribing for viral respiratory infections. Clin. Infect. Dis. 29: 155–160.
123. Weinberg, J; Grimaud, O; Newton, L. (1999). Establishing priorities for European collaboration in communicable disease surveillance. Eur. J. Public Health. 9(3): 236–240.
124. Weir, M; Rajić, A; Ditul, L; Uhland, C; Bruneau, N. (2012). Zoonotic bacteria and antimicrobial resistance in aquaculture: Opportunities for surveillance in Canada. Can. Vet. J. 59: 619–622.
125. Weiss, K; Blais, R; Forint, A; Lantin, S; Gaudet, M. (2011). Impact of a multipronged education strategy on antibiotic prescribing in Quebec, Canada. Clin. Infect. Dis. 53: 433–439.
126. White, AR; BSAC Working Parties on Resistance Surveillance. (2008). The British Society for Antimicrobial Chemotherapy resistance surveillance project: A successful collaborative model. J. Antimicrob. Chemother. 62(Suppl 2): 3–14.
127. Wilson, J; Conly, J; Wong, T; Jayaraman, G; Sargeant, J; Papadopoulos, A; Young, V; Quist-Moyer, M; Bauer, S. (2010). Strategies to control community-associated antimicrobial resistance among enteric bacteria and methicillin-resistant Staphylococcus aureus in Canada – Executive summary. Can. J. Infect. Dis. Med. Microbiol. 21(3): 133–137.
128. Wray, C; Gnanou, JC. (2000). Antibiotic resistance monitoring in bacteria of animal origin: Analysis of national monitoring programmes. Int. J. Antimicrob. Agents. 14(4): 291–294.
129. Zoutman, DE; Ford, BD. (2005). The relationship between hospital infection surveillance and control activities and antibiotic-resistant pathogen rates. Am. J. Infect. Control. 33: 1–5.
Appendix C.
Survey Questionnaire
This survey represents part of a National Collaborating Centre for Infectious Diseases funded project, with the ultimate aim of developing recommendations for an optimized, comprehensive Canadian program of antimicrobial use and resistance surveillance. Our data collection phase involves a systematic literature review of available use and resistance surveillance systems, and this questionnaire which we are administering to key informants in public health, microbiology and biology, and human and veterinary health. The goal of this questionnaire is to identify “what is missing” in the areas of antimicrobial use and resistance surveillance. All information you provide is confidential and will be used for the purposes of this research project only.
For more detailed information about the project or if you have any questions, please contact Dr. Patricia Keen at plkeenpl@civil.ubc.ca.
Project Team Leaders:
Dr. Lynora Saxinger, University of Alberta
Dr. Jennifer Grant, Vancouver General Hospital
Dr. David Patrick, University of British Columbia
Project Team:
Dr. Patricia Keen, University of British Columbia
Ms. Diana Kao, University of British Columbia
Your participation in this interview implies consent to provide information for our study. You may stop this interview and survey at any time.
Please describe your role in public health, antimicrobial use or resistance work:
1.
Name:
M / F
Profession/discipline:
Years of practice:
Specialization:
Role(s) pertaining to Antimicrobial Use and Resistance:
Province/country:
2. To understand your interactions with these types of data, please describe:
a. What information do you access about antibiotic use and antibiotic resistance in your patient population? (either human or veterinary)
b. How do you access this information?
c. How do you use this information?
d. What information, that you view as valuable, about antibiotic use do you feel is currently missing or inaccessible?
e. What information, that you view as valuable, about antibiotic resistance do you feel is currently missing or inaccessible?
f. How would you like to access information?
g. If you are a holder of data, what barriers do you see to sharing data for the purposes of reporting and how do you think these barriers could be overcome?
3. Do you think that:
a. Surveillance of antimicrobial use and antimicrobial resistance in human medicine is a reasonable priority on which we should spend resources? Why or why not?
b. Surveillance of antimicrobial use and antimicrobial resistance in veterinary and agri-food settings is a reasonable priority on which we should spend resources? Why or why not?
4. What Canada wide, other country wide, or international systems are you aware of (or do you participate in) that collect data in the area of:
a. Human antimicrobial use (Canadian or international)
b. Antimicrobial resistance in human pathogens (Canadian or international)
c. Veterinary and agri-food antimicrobial use (Canadian or international)
d. Antimicrobial resistance in veterinary pathogens (Canadian or international)
Of those, do you feel any should be highlighted as an exemplary data collection system? Why?
5. What do you think are the KEY elements of a human/veterinary antimicrobial USE monitoring system?
6. What do you think are the KEY elements of a human/veterinary antimicrobial RESISTANCE monitoring system?
7. For questions 5 & 6 (use and resistance surveillance) please prioritize (1-4) the following “key elements” of resistance and utilization in Canada
• Timely (give reporting frequency and maximum delay for reports)
• Comprehensive (putting use, resistance, human and veterinary together)
• Coordinated (Done provincially, nationally or with identified priorities across populations)
• Making raw data available for assessment by interested groups• Issuing reports with analysis included
• Actionable (reported to a group that has responsibility and oversight)
8. Please rate Canadian surveillance systems based on information you encounter through your work in relevant fields. The ratings will be on a scale of 10, with one representing “poor or nonexistent” surveillance systems and 10 representing “excellent” surveillance. Please indicate if you do not know or prefer not to give an opinion.
a. How would you rate Canadian human antibiotic USE surveillance? Please comment on why you gave this rating.
b. How would you rate Canadian human antibiotic RESISTANCE surveillance? Please comment on why you gave this rating.
c. How would you rate Canadian VETERINARY and AGRI-FOOD antimicrobial USE surveillance? Please comment on why you gave this rating.
d. How would you rate Canadian VETERINARY antimicrobial resistance surveillance? Please comment on why you gave this rating.
9. In your opinion, who should be responsible (e.g. government (which level or ministry?), professional group etc.) for the collection and dissemination of antimicrobial use/resistance data?
10. In your opinion, should there be restrictions on who has access to data or should collected data be made fully public? If restricted, to whom should it be restricted?
11. Are there priority organisms for surveillance? If yes, please specify which organisms you think are most important.
12. Are there priority drugs for surveillance? If yes, please specify?
13. Do you think any specific human populations should be a priority for surveillance of antimicrobial use? Of antimicrobial resistance? (e.g. specific hospital units, specific infection risk group, specific exposures) (allow not to answer if not familiar with human medicine).
a. If yes, what populations?
b. Why?
14. Do you think specific veterinary populations should be a priority for surveillance of antimicrobial use? Of antimicrobial resistance? (e.g. poultry, swine etc.)? (Allow not to answer if not familiar with veterinary medicine and/or agri-food). a. If yes, what populations? b. Why?
15. Should there be reporting of antimicrobial utilization in human medicine? In veterinary medicine?
a. If yes – should reporting be mandatory or voluntary? Why?
b. Should reporting, either mandatory or voluntary, depend on the indication of the prescription or regardless of purpose of the prescription?
16. Do you think veterinary and human antimicrobial utilization data should be reported together or separately? What about antimicrobial resistance data?
17. Specifically in veterinary antimicrobial use/resistance surveillance, do you feel a distinction should be made between antimicrobial use and resistance in the context of growth promotion, prophylactic therapy or treatment of illness?
a. If you think there is a distinction to be made, please provide your rationale.
18. We are asking our participants to identify two other individuals who might be able to provide additional perspective on human and animal antimicrobial resistance and antimicrobial utilization monitoring in Canada. Please can you suggest two colleagues who may also be willing to answer these questions?
Thank you for your participation in this project.
Appendix D.
Survey and Results
A total of 272 experts were invited to participate in the survey interview process between January 2 and May 10, 2013 with 147 completing the survey questionnaire yielding a response rate of 53%. Figure 1 represents the experts from various domains in human medicine who were contacted and who completed the survey, and Figure 2 shows the same data for the animal health community.

Figure 1: Number of human medical professionals who completed the survey interview relative to the total number of individuals invited to participate in the survey.

Figure 2: Number of veterinary medical professionals who completed the survey interview relative to the total number of individuals invited to participate in the study.

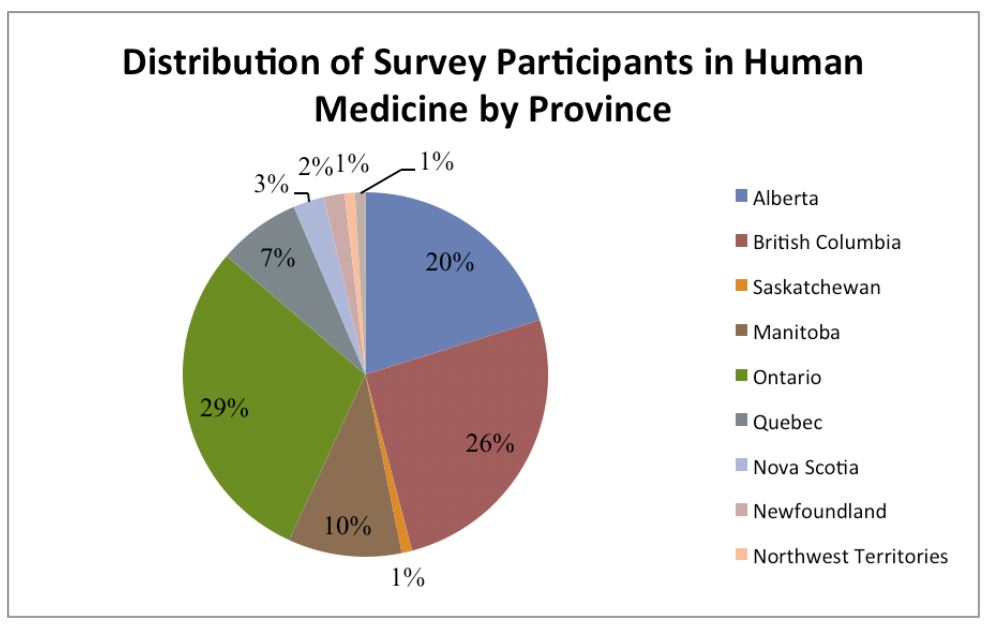

Figure 3: Distribution of survey participants by province
Key informants representing a broad range of sectors and disciplines with interest in the health consequences of use of antibiotics and development of antimicrobial resistance in pathogens in both human medicine and veterinary medicine were invited to participate in the survey via in-person or telephone interviews. The total number of survey participants was 146 (104 human medical professionals and 42 veterinary medical professionals) and the overall survey response rate was 56%. Regional distribution of the survey respondents are illustrated in Figures G.1–G.3 with distribution by province presented in Appendix D. Some key respondents represented North American or global organizations with offices in the United States but their professional role required an involvement in issues related to antibiotic use or antimicrobial resistance surveillance in Canada.
The survey instrument is reproduced in Appendix B.
Selected results of the data are summarized here, and a more comprehensive qualitative data summary will become available in a separate report.
When asked whether antibiotic use data or AMR data should be reported together or separately, the respondents from veterinary and agri-food sectors were more in favour of integrated reporting.
| Respondents: Human Health | Respondents: Veterinary/Agri-food | Total Respondents | |
|---|---|---|---|
| Data should be Reported Together | 25% | 64% | 37% |
| Data should be reported Separately | 63% | 19% | 50% |
A majority of the respondent from both sectors were in favour of mandatory reporting over voluntary reporting. Survey participants were asked whether reporting in human medicine should be voluntary or mandatory:
| Respondents: Human Health | Respondents: Veterinary/Agri-food | Total Respondents | |
|---|---|---|---|
| Mandatory Reporting | 64% | 71% | 66% |
| Voluntary Reporting | 17% | 2% | 13% |
The same question in veterinary reporting had a similar distribution (79% human medicine respondents and 71% veterinary respondents) of those in favour of mandatory reporting.
Participants were asked should if reporting on antimicrobial utilization in human medicine, either mandatory or voluntary, should depend on the indication of the prescription or regardless of purpose of the prescription, and respondents (69% human medicine respondents and 71% veterinary-agri-food respondents) were in favour of reporting regardless of prescription indication. An even higher proportion were in favour of reporting regardless of prescription indication in veterinary medicine (75% and 76& respectively.)
When asked which institutions should be responsible for the collection and dissemination of antimicrobial use/resistance data, respondents expressed different opinions regarding partnerships between federal government, provincial government and professional groups: Sixty-seven percent of human medicine respondents identified the federal government plus or minus provincial governments and professional groups as the responsible group, and 13% and 14% identified provincial governments and professional groups as the sole responsible groups respectively. Ninety-one percent of veterinary and agri-food respondents identified the federal government plus or minus provincial governments and professional groups as the responsible group, with a larger proportion identified a federal-provincial and industry collaboration, and only 2% identifying the provincial governments as responsible.
| Responsibility for AMR-AMU surveillance | Respondents: Human Health (%) | Respondents: Veterinary/Agri-food (%) | Total Respondents (%) |
|---|---|---|---|
| Federal | 35 | 33 | 34 |
| Federal plus Provincial | 28 | 36 | 30 |
| Federal plus professional group | 3 | 10 | 5 |
| Federal Provincial And Industry | 1 | 12 | 4 |
| Professional Groups | 14 | 0 | 10 |
| Provincial | 10 | 2 | 10 |
Respondents were asked whether there should be restrictions on who has access to surveillance data or should the collected data be made fully public, with a fairly even proportion in each sectors in favour of fully public and partially restricted data.
| Surveillance Data Availability | Respondents: Human Health | Respondents: Veterinary/Agri-food | Total Respondents |
|---|---|---|---|
| Fully Public | 49% | 48% | 49% |
| Some Restrictions | 51% | 52 | 51% |
Figure 4: Regional distribution of total survey respondents
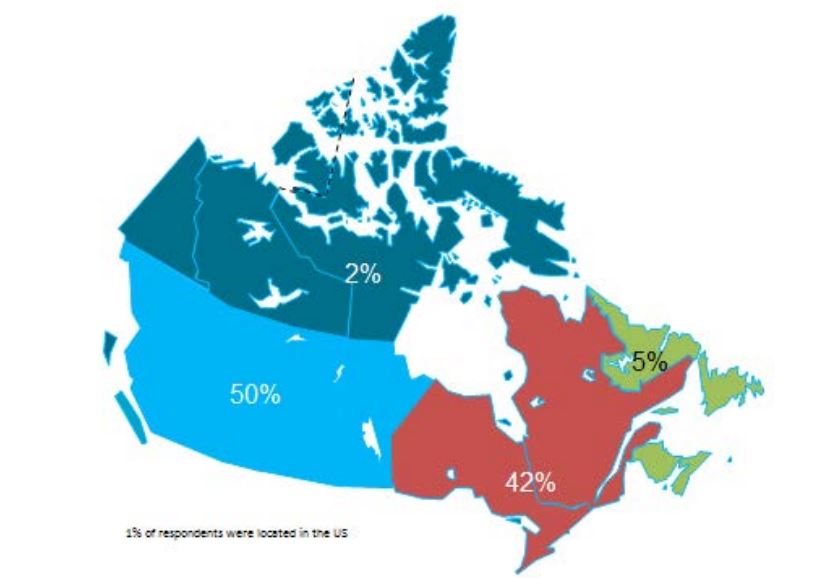
Figure 5: Regional distribution of human medicine related respondents
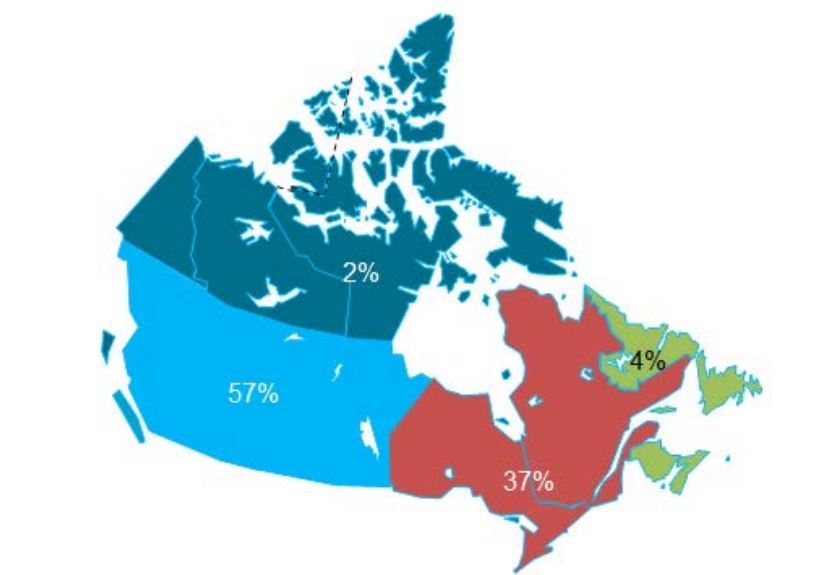
Figure 6: Regional distribution of veterinary medicine related respondents
Appendix E.
Recommendations of Previous Canadian AMR Conferences and Proceedings
This section contains excerpts and summaries from a variety of previous reports, conferences, and policy papers, sometimes renumbered for ease of reference.
1. Controlling Antimicrobial Resistance: An Integrated Action Plan for Canadians Health Canada and Canadian Infectious Disease Society, 1997
This report arose from a large national Consensus Conference in Montreal in 1997. The following recommendations are provided verbatim from the original document since they remain highly relevant to antimicrobial stewardship.
1.1. To identify structures and key human resources at the care-setting and (local) regional levels that are/will be most responsible for coordinating the care of clients/patients/consumers affected by antimicrobial resistant organisms.
1.2. To improve funding and access to expert resources on antibiotic use in all Canadian health care settings. This will be accomplished by the creation of expert panels to promote local antibiotic-use protocols and to provide case consultations as an adjunct to existing provincial/territorial or regional public health networks.
1.3. To establish antibiotic stewardship and antibiotic use teams in all Canadian hospitals by: a. incorporating them into accreditation standards; b. obtaining support from the medical and administrative leadership.
1.4. To establish antimicrobial usage, monitoring, and intervention programs at the long-term care institutional level.
1.4.1. Short term: monitoring of antimicrobial usage
1.4.2. Intermediate term: monitoring antimicrobial appropriateness
1.4.3. Long term: optimizing antimicrobial use
1.5. To reduce overall antimicrobial usage (prescriptions) by 25% within 3 years by focusing on community-acquired respiratory infection.
2. National Action Plan to Address Antibiotic Resistance, Canadian Committee on Antibiotic Resistance (CCAR), 2004
This report reflected on the lack of progress since the initial meetings in 1997 and included recommendations on surveillance in the context of an AMR response, excerpted below. A summary of actions identified in the four key areas of surveillance, infection prevention and control, optimal use and research have been identified … a number of these actions include the revitalization of earlier initiatives that were overtaken by other public health issues … During 2004, CCAR will actively solicit the endorsement of this Action Plan by all of the key organizations that must take a leadership role as well as those responsible for the Action Plan implementation.
Surveillance:
2.1. Current surveillance systems, including CNISP and CIPARS, will be expanded to include a wider variety of facilities and organisms. Health Canada, the Canadian Hospital Epidemiology Committee and CCAR will consider a pilot project for a new real time surveillance system to monitor resistance patterns in one key area of interest by the end of 2004.
2.2. In 2005, Health Canada, provincial Ministries of Health, the Canadian Hospital Epidemiology Committee, CCAR and IMS Health will discuss mechanisms to collect, analyze and compare antibiotic use data from human health care facilities, retail pharmacies and other private and public databases that are available.
2.3. Health Canada and key provincial Ministries of Health and Agriculture, in conjunction with CCAR and other stakeholders, will form or revitalize Steering Committees on Surveillance in 2004 to escalate current efforts to monitor antibiotic use and resistance in human health and agri-food settings.
2.4. CEQA-AGAR, with support from Health Canada and provincial laboratories, will restart their efforts by the end of 2005to ensure existing and emerging resistance is monitored and that laboratory methodologies are standardized.
3. Obstacles to Developing a Multinational Report Card on Antimicrobial Resistance for Canada: An Evidence Based Review (Stephens et al., Microbial Drug Resistance 13: 4, 251-259), 2007
This publication reviewed the possibility of comparing Canada’s AMR data to other countries- basically, developing a “report card” to allow comparisons between countries AMR programs. It is included, even though it was a literature search/grey literature piece, because it extensively reviewed Canadian programs.
Commentary pertinent to Canadian AMR surveillance included:
3.1. Canadian program objectives have not been precisely articulated, and measurable performance objectives are lacking in most AMR programs.
3.2. Information for Canadian programs was dispersed through various locations (web sites, peer reviewed press, institutional memory). Whereas many individual programs provided useful data on specific issues for specific times and locations, differences in methods, funding and infrastructure created obstacles to integrating all results into a single comprehensive national picture.
3.3. The lack of explicit thresholds or targets for success in Canadian and most international AMR programs made it impossible to determine if a country was more or less effective in meeting their goals when compared with Canada
3.4. Programs developed for local or provincial planning tended not to be connected with similar programs in other Canadian jurisdictions. The lack of coordination was a fundamental obstacle to developing a comprehensive, ongoing national picture.
3.5. The most common method for disseminating AMR results outside a jurisdiction was through the scientific press or at scientific meetings (with limited access to full text reports and significant time lags).
3.6. Canada did not have a single comprehensive program for collecting and integrating data on AMR, drug use, and infection control, resulting in a patchwork of projects with varying methodologies (many with unexamined potential for significant selection and sampling bias), interpretation, sustainability, finding and objectives that are not effectively knit together.
4. Pan Canadian Stakeholder Consultations on Antimicrobial Resistance, 2010
Prior to the disbanding of the Canadian Committee on Antibiotic Resistance (CCAR) group, the AMR Consultation process was held, and the findings summarized in a report in September 2009. Noteworthy excerpts are below, including a discussion of the barriers to implementation faced after the 2004 CCAR report, and a summary of the recommendations.
4.1. CHALLENGES
…a number of specific challenges were faced by the CCAR as it sought to meet its mandate:
4.1.1. There was not an adequately staffed infrastructure (i.e. secretariat) to coordinate and/or integrate AMR activities nationally and there was a lack of fulltime employees to assist with implementation. Historically, when actions were identified they were implemented on a voluntary basis by the CCAR Board and the community of practice at large;
4.1.2. There was no identified lead for AMR within the federal government. The current link between the CCAR and the federal government was not at an appropriate level to move policy items ahead and ensure AMR issues were being heard by senior officials within government; and
4.1.3. The funding provided to the CCAR was not sufficient given the breadth of responsibilities falling under its mandate. There was little additional funding directed towards the National Action Plan, let alone new and emerging priorities.
4.2. SURVEILLANCE: There is a need to appoint a lead to oversee AMR surveillance activities across Canada. The lead organization should develop an AMR Surveillance Working Group and oversee the development and implementation of a Pan-Canadian AMR Surveillance Plan that integrates surveillance activities across the multiple sectors (i.e. animal, human, environmental) and is standardized, timely, easily accessible and responsive to its multiple users (e.g. local, rural, laboratories, pharmacies, governments, etc) and the Canadian public at large.
4.3. ANTIMICROBIAL STEWARDSHIP: It was determined that Canada needs to establish a lead for antimicrobial stewardship who – in partnership with key stake- holders – will oversee the development and implementation of a comprehensive Pan-Canadian Antimicrobial Stewardship Plan. Stakeholders require, among other things, easy access to antimicrobial usage guidelines and surveillance data on antimicrobial utilization and antimicrobial resistant organisms. As well, enforcement of appropriate antimicrobial use in both the human and animal sectors needs to be coordinated.
4.4. EDUCATION and TRAINING: Canada must build on existing AMR education/training campaigns, combine education/training with other strategies such as regulation, and support increased collaboration between schools and institutions. There was strong support for a rollout of the Do Bugs Need Drugs? (DBND) campaign and the Bugs & Drugs antimicrobial reference guide on a national scale.
4.5. GOVERNANCE: The participants encouraged the Public Health Agency of Canada to take the lead in moving AMR forward within the federal government of Canada. Several common themes were identified that should be incorporated into the new governance model for AMR. The themes include:
4.5.1. Secure funding from multiple agencies/government departments;
4.5.2. Develop a secretariat/coordinating body responsible for overall coordination & integration;
4.5.3. Link to a high level governmental decision making body; and
4.5.4. Build the governance model around existing successful AMR activities and existing action plans.
5. British Columbia Public Health Office (British Columbia PHO, 2000).
In 2000, the B.C. PHO issued a statement summarizing a series of actions specific to surveillance efforts to manage human health risks associated with antimicrobial resistance. This was the only specific province statement that was found dealing with AMR. The planned actions included:
5.1. Developing a coordinated provincial plan for surveillance of antibiotic use and antimicrobial resistance in human and non-human settings.
5.2. Identifying and coordinating source data related to antimicrobial resistance and associated impacts on morbidity and mortality.
5.3. Defining and standardizing definitions and methods for surveillance data.
5.4. Developing integrated computer systems and database networks.
5.5. Developing platforms for the dissemination of information and the publication of regular reports.
6. Canadian Medical Association Annual Meeting Resolutions (2010)
Since 2010, the resolutions approved at the annual meeting of the CMA include specific recommendations related to addressing the issue of antimicrobial resistance in agriculture and veterinary medicine in Canada.
The approved CMA resolutions that pertain to antibiotics in animals are:
6.1. The Canadian Medical Association recommends that the Food and Drugs Act and its regulations be amended to close the “own use” provision for the unmanaged importation of antibiotics for agricultural use.
6.2. The Canadian Medical Association supports the development of a national system to identify and report the identities and quantities of antibiotics acquired domestically or imported for use in food animals.
6.3. The Canadian Medical Association supports regulations to severely limit the use of medically important antibiotics on animals being raised for human consumption.
6.4. The Canadian Medical Association recommends that a prescription from a veterinarian be required for all antibiotics used in the raising of farm animals or for any other agricultural purpose.
6.5. The Canadian Medical Association calls on the federal government to investigate animal husbandry techniques that decrease the need for antibiotics in animals and to support techniques proven to be effective.
6.6. The Canadian Medical Association, in collaboration with provincial/territorial medical associations, will work with Health Canada and the Public Health Agency of Canada to investigate the agriculture-related release of antibiotic resistant organisms and residual antibiotics into earth and water ecosystems, as well as the role they play in the emergence of antibiotic resistant organisms in humans.
6.7. The Canadian Medical Association will encourage Health Canada and Agriculture and Agri-Food Canada to work with other relevant government or non-government agencies to develop a comprehensive national strategy to combat antimicrobial resistance.
7. Policy Paper: When Antibiotics Stop Working, Ontario Medical Association, 2013
Recommendations in the report included: encouraging the federal government to provide funding for research, strengthened surveillance, and educational campaigns focused on antibiotic resistance, to promote the use of electronic records to allow physicians to compare patients’ past prescriptions and diagnoses, to close the ‘own-use loophole’ of antibiotic use in agriculture, and to establish surveillance in areas where it does not exist (agriculture) and strengthened in areas where it does exist (medicine) in order to collect data and gain a firmer understanding about antibiotic resistance in both humans and animals.
Appendix F.
Categorization of antimicrobial drugs based on importance in human medicine (CIPARS, 2008)
I: Very High Importance
Antimicrobial class
Carbapenems
Cephalosporins (the 3rd and 4th generations)
Fluoroquinolones
Glycopeptides
Glycylcyclines
Ketolides
Lipopeptides
Monobactams
Nitroimidazoles (metronidazole)
Oxazolidinones
Penicillin-β-lactamase inhibitor combinations
Polymyxins (colistin)
Therapeutic agents for tuberculosis (e.g. ethambutol, isoniazid, pyrazinamide, and rifampin)
II: High Importance
Antimicrobial class
Aminoglycosides (except topical agents)
Cephalosporins (the first and second generations: including cephamycins)
Fusidic acid; Lincosamides
Macrolides
Penicillins
Quinolones (except fluoroquinolones)
Streptogramins
Trimethoprim-sulfamethoxazole
III: Medium Importance
Antimicrobial class
Aminocyclitols
Aminoglycosides (topical agents)
Bacitracins
Fosfomycin
Nitrofurans
Phenicols
Sulfonamides
Tetracyclines
Trimethoprim
IV: Low Importance
Antimicrobial class
Flavophospholipols
Ionophores